Intratumoral STING agonist reverses immune evasion in PD-(L)1-refractory Merkel cell carcinoma: mechanistic insights from detailed biomarker analyses
- PMID: 39401968
- PMCID: PMC11474899
- DOI: 10.1136/jitc-2024-009803
Intratumoral STING agonist reverses immune evasion in PD-(L)1-refractory Merkel cell carcinoma: mechanistic insights from detailed biomarker analyses
Abstract
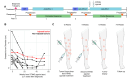
Background: Antibodies blocking programmed death (PD)-1 or its ligand (PD-L1) have revolutionized cancer care, but many patients do not experience durable benefits. Novel treatments to stimulate antitumor immunity are needed in the PD-(L)1 refractory setting. The stimulator of interferon genes (STING) protein, an innate sensor of cytoplasmic DNA, is a promising target with several agonists in development. However, response rates in most recent clinical trials have been low and mechanisms of response remain unclear. We report detailed biomarker analyses in a patient with anti-PD-L1 refractory, Merkel cell polyomavirus (MCPyV)-positive, metastatic Merkel cell carcinoma (MCC) who was treated with an intratumoral (IT) STING agonist (ADU-S100) plus intravenous anti-PD-1 antibody (spartalizumab) and experienced a durable objective response with regression of both injected and non-injected lesions.
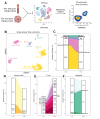
Methods: We analyzed pretreatment and post-treatment tumor and peripheral blood samples from our patient with single-cell RNA sequencing, 30-parameter flow cytometry, T cell receptor sequencing, and multiplexed immunohistochemistry. We analyzed cancer-specific CD8 T cells using human leukocyte antigen (HLA)-I tetramers loaded with MCPyV peptides. We also analyzed STING expression and signaling in the tumor microenvironment (TME) of 88 additional MCC tumor specimens and in MCC cell lines.
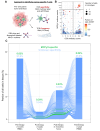
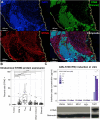
Results: We observed high levels of MCPyV-specific T cells (12% of T cells) in our patient's tumor at baseline. These cancer-specific CD8 T cells exhibited characteristics of exhaustion including high TOX and low TCF1 proteins. Following treatment with STING-agonist plus anti-PD-1, IT CD8 T cells expanded threefold. We also observed evidence of likely improved antigen presentation in the MCC TME (greater than fourfold increase of HLA-I-positive cancer cells). STING expression was not detected in any cancer cells within our patient's tumor or in 88 other MCC tumors, however high STING expression was observed in immune and stromal cells within all 89 MCC tumors.
Conclusions: Our results suggest that STING agonists may be able to work indirectly in MCC via signaling through immune and stromal cells in the TME, and may not necessarily need STING expression in the cancer cells. This approach may be particularly effective in tumors that are already infiltrated by inflammatory cells in the TME but are evading immune detection via HLA-I downregulation.
Keywords: Abscopal; Intralesional; Pathogen-Associated Molecular Pattern - PAMP; Skin Cancer; T cell.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: PN reports personal fees from Pfizer, Bristol Myers Squibb, EMD Serono, Rain Therapeutics, Almirall, and Instill Bio. In addition, PN and DK have pending patents for “Merkel cell polyomavirus T antigen-specific TCRs and uses thereof” and “Novel epitopes as T cell targets in Merkel Cell Carcinoma (MCC)”. CC has a pending patent “Merkel cell polyomavirus T antigen-specific TCRs and uses thereof”. LT reports research grant funding (to institution) from Seagen and Merck. SB has received consulting or advisory role fees and honoraria from Bristol Myers Squibb, Sanofi/Regeneron, and Incyte; and has received research funding from Bristol Myers Squibb, Merck, EMD Serono, Exicure, Incyte, Checkmate Pharmaceuticals, 4SC, Seven and Eight Pharmaceuticals, Amphivena Therapeutics, TriSalus Life Sciences, Novartis and Agenus. DK reports research funding from Sanofi Pasteur. DK has received consulting or advisory fees from Curevo Vaccine and MaxHealth LLC Pasteur. KS has received consulting or advisory role fees from Sensei Biotherapeutics.
Figures


References
-
- Downey CM, Aghaei M, Schwendener RA, et al. DMXAA causes tumor site-specific vascular disruption in murine non-small cell lung cancer, and like the endogenous non-canonical cyclic dinucleotide STING agonist, 2’3’-cGAMP, induces M2 macrophage repolarization. PLoS ONE. 2014;9:e99988. doi: 10.1371/journal.pone.0099988. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
