Viral entry mechanisms: the role of molecular simulation in unlocking a key step in viral infections
- PMID: 39402013
- PMCID: PMC11788750
- DOI: 10.1002/2211-5463.13908
Viral entry mechanisms: the role of molecular simulation in unlocking a key step in viral infections
Abstract
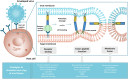
Viral infections are a major global health concern, affecting millions of people each year. Viral entry is one of the crucial stages in the infection process, but its details remain elusive. Enveloped viruses are enclosed by a lipid membrane that protects their genetic material and these viruses are linked to various human illnesses, including influenza, and COVID-19. Due to the advancements made in the field of molecular simulation, significant progress has been made in unraveling the dynamic processes involved in viral entry of enveloped viruses. Simulation studies have provided deep insight into the function of the proteins responsible for attaching to the host receptors and promoting membrane fusion (fusion proteins), deciphering interactions between these proteins and receptors, and shedding light on the functional significance of key regions, such as the fusion peptide. These studies have already significantly contributed to our understanding of this critical aspect of viral infection and assisted the development of effective strategies to combat viral diseases and improve global health. This review focuses on the vital role of fusion proteins in facilitating the entry process of enveloped viruses and highlights the contributions of molecular simulation studies to uncover the molecular details underlying their mechanisms of action.
Keywords: enveloped viruses; fusion peptide; membrane fusion; molecular simulation; receptor binding; viral entry.
© 2024 The Author(s). FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
[Entry process of enveloped viruses to host cells].Uirusu. 2009 Dec;59(2):205-13. doi: 10.2222/jsv.59.205. Uirusu. 2009. PMID: 20218329 Review. Japanese.
-
Roles of Cholesterol in Early and Late Steps of the Nipah Virus Membrane Fusion Cascade.J Virol. 2021 Feb 24;95(6):e02323-20. doi: 10.1128/JVI.02323-20. Print 2021 Feb 24. J Virol. 2021. PMID: 33408170 Free PMC article.
-
Viral Membrane Fusion and the Transmembrane Domain.Viruses. 2020 Jun 27;12(7):693. doi: 10.3390/v12070693. Viruses. 2020. PMID: 32604992 Free PMC article. Review.
-
New Biophysical Approaches Reveal the Dynamics and Mechanics of Type I Viral Fusion Machinery and Their Interplay with Membranes.Viruses. 2020 Apr 8;12(4):413. doi: 10.3390/v12040413. Viruses. 2020. PMID: 32276357 Free PMC article. Review.
-
Entry of Enveloped Viruses into Host Cells: Membrane Fusion.Subcell Biochem. 2024;105:567-592. doi: 10.1007/978-3-031-65187-8_16. Subcell Biochem. 2024. PMID: 39738958 Review.
Cited by
-
Challenges in simulating whole virus particles and how to fix them with the SIRAH force field.Biophys Rev. 2025 Mar 22;17(2):285-292. doi: 10.1007/s12551-025-01305-x. eCollection 2025 Apr. Biophys Rev. 2025. PMID: 40376428 Review.
-
Structural Bioinformatics: exciting times in a rapidly evolving field.FEBS Open Bio. 2025 Feb;15(2):200-201. doi: 10.1002/2211-5463.13968. FEBS Open Bio. 2025. PMID: 39895624 Free PMC article.
References
-
- Morens DM and Al E (2020) The COVID‐19 pandemic: what can we learn from it? J Clin Invest 130, 2140–2145.
-
- Simons E, Ferrari M, Fricks J, Wannemuehler K, Anand A, Burton A and Strebel P (2012) Assessment of the 2010 global measles mortality reduction goal: results from a model of surveillance data. Lancet 379, 2173–2178. - PubMed
-
- Fields BN, Knipe DM, Howley PM and Griffin DE (2007) Fields' Virology. Lippincott Williams & Wilkins.
-
- Patel MM and Al E (2008) Global epidemiology of hepatitis a virus infection: new estimates of age‐specific antibody to HAV seroprevalence. Hepatology 48, 1333–1341. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

