RNA architecture of porcine deltacoronavirus genome inside virions detected by vRIC-seq
- PMID: 39402053
- PMCID: PMC11473776
- DOI: 10.1038/s41597-024-03975-w
RNA architecture of porcine deltacoronavirus genome inside virions detected by vRIC-seq
Abstract
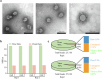
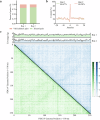
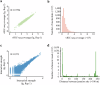
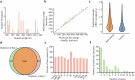
Porcine deltacoronavirus (PDCoV) is a newly emerging and special delta coronavirus, which infect mammals such as pigs, cattle and humans, as well as chickens and birds. Exploring RNA structures in the viral genome benefits the understanding of the role of RNA in the lifecycle of viruses. In this study, vRIC-seq is employed to analyze the RNA-RNA interaction in the whole genome structure of PDCoV in virions. About 12.87 and 13.52 million paired reads are obtained in two biological replicates, respectively, with 17.9% and 14.8% of them are identified as valid chimeric reads. These are employed to predict the RNA secondary structure, which is compact and highly structured. A twisted-cyclized conformation is observed in the RNA-RNA interaction map of PDCoV for the first time. 77 multi-way junctions are evenly distributed in the PDCoV genome. Our work provides fundamental structural insights that are essential for understanding the genomic structure and function, genetic evolution, and packaging characteristics of PDCoV.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

