Mycobacterium avium subspecies paratuberculosis (MAP) infection, and its impact on gut microbiome of individuals with multiple sclerosis
- PMID: 39402079
- PMCID: PMC11479286
- DOI: 10.1038/s41598-024-74975-4
Mycobacterium avium subspecies paratuberculosis (MAP) infection, and its impact on gut microbiome of individuals with multiple sclerosis
Abstract
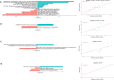
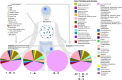
The microbial ecology of Mycobacterium avium subspecies paratuberculosis infections (MAP) within the context of Multiple Sclerosis (MS) is largely an unexplored topic in the literature. Thus, we have characterized the compositional and predicted functional differences of the gut microbiome between MS patients with MAP (MAP+) and without (MAP-) infection. This was done in the context of exposome differences (through self-reported filled questionnaires), principally in anthropometric and sociodemographic patterns to gain an understanding of the gut microbiome dynamics. 16S rRNA microbiome profiling of faecal samples (n = 69) was performed for four groups, which differed by disease and MAP infection: healthy cohort (HC) MAP-; HC MAP+ ; MS MAP-; and MS MAP+ . Using a dynamic strategy, with MAP infection and time of sampling as occupancy models, we have recovered the core microbiome for both HC and MS individuals. Additional application of neutral modeling suggests key genera that are under selection pressure by the hosts. These include members of the phyla Actinobacteriota, Bacteroidota, and Firmicutes. As several subjects provided multiple samples, a Quasi Conditional Association Test that incorporates paired-nature of samples found major differences in Archaea. To consolidate treatment groups, confounders, microbiome, and the disease outcome parameters, a mediation analysis is performed for MS cohort. This highlighted certain genera i.e., Sutterella, Akkermansia, Bacteriodes, Gastranaerophilales, Alistipes, Balutia, Faecalibacterium, Lachnospiraceae, Anaerostipes, Ruminococcaceae, Eggerthellaceae and Clostridia-UCG-014 having mediatory effect using disease duration as an outcome and MAP infection as a treatment group. Our analyses indicate that the gut microbiome may be an important target for dietary and lifestyle intervention in MS patients with and without MAP infection.
Keywords: Gut microbiome; 16S rRNA; Mediation analysis; Neutral modelling; Multiple sclerosis.
© 2024. The Author(s).
Conflict of interest statement
PD is employed by BIOMES NGS GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Faecal microbial diversity in a cattle herd infected by Mycobacterium avium subsp. paratuberculosis: a possible effect of production status.World J Microbiol Biotechnol. 2024 Jul 22;40(9):276. doi: 10.1007/s11274-024-04080-1. World J Microbiol Biotechnol. 2024. PMID: 39037634 Free PMC article.
-
The Impact of Mycobacterium avium subsp. paratuberculosis on Intestinal Microbial Community Composition and Diversity in Small-Tail Han Sheep.Pathogens. 2024 Dec 18;13(12):1118. doi: 10.3390/pathogens13121118. Pathogens. 2024. PMID: 39770377 Free PMC article.
-
A Machine Learning Approach Reveals a Microbiota Signature for Infection with Mycobacterium avium subsp. paratuberculosis in Cattle.Microbiol Spectr. 2023 Feb 14;11(1):e0313422. doi: 10.1128/spectrum.03134-22. Epub 2023 Jan 19. Microbiol Spectr. 2023. PMID: 36656029 Free PMC article.
-
Potential application of emerging diagnostic techniques to the diagnosis of bovine Johne's disease (paratuberculosis).Vet J. 2016 Mar;209:32-9. doi: 10.1016/j.tvjl.2015.10.033. Epub 2015 Oct 22. Vet J. 2016. PMID: 26831164 Review.
-
Molecular Epidemiology of Mycobacterium avium subsp. paratuberculosis on Dairy Farms.Annu Rev Anim Biosci. 2016;4:155-76. doi: 10.1146/annurev-animal-021815-111304. Epub 2015 Nov 2. Annu Rev Anim Biosci. 2016. PMID: 26526547 Review.
References
-
- WHO. Multiple sclerosis: World Health Organization. https://www.who.int/news-room/fact-sheets/detail/multiple-sclerosis (2023).
-
- Olsson, T., Barcellos, L. F. & Alfredsson, L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat. Rev. Neurol. 13(1), 25–36 (2017). - PubMed
-
- Ekundayo, T. C. et al. Systematic review and meta-analysis of Mycobacterium avium subsp. paratuberculosis as environmental trigger of multiple sclerosis. Mult. Scler. Relat. Disord. 59, 103671 (2022). - PubMed
-
- Hayashi, F. et al. Elevated mycobacterium avium subsp paratuberculosis (MAP) antibody titer in Japanese multiple sclerosis. J. Neuroimmunol. 360, 577701 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

