Unraveling autophagic imbalances and therapeutic insights in Mecp2-deficient models
- PMID: 39402139
- PMCID: PMC11555085
- DOI: 10.1038/s44321-024-00151-w
Unraveling autophagic imbalances and therapeutic insights in Mecp2-deficient models
Abstract
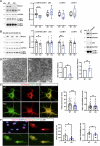
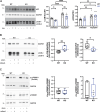
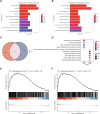
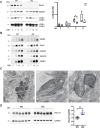
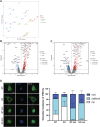
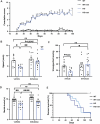
Loss-of-function mutations in MECP2 are associated to Rett syndrome (RTT), a severe neurodevelopmental disease. Mainly working as a transcriptional regulator, MeCP2 absence leads to gene expression perturbations resulting in deficits of synaptic function and neuronal activity. In addition, RTT patients and mouse models suffer from a complex metabolic syndrome, suggesting that related cellular pathways might contribute to neuropathogenesis. Along this line, autophagy is critical in sustaining developing neuron homeostasis by breaking down dysfunctional proteins, lipids, and organelles.Here, we investigated the autophagic pathway in RTT and found reduced content of autophagic vacuoles in Mecp2 knock-out neurons. This correlates with defective lipidation of LC3B, probably caused by a deficiency of the autophagic membrane lipid phosphatidylethanolamine. The administration of the autophagy inducer trehalose recovers LC3B lipidation, autophagosomes content in knock-out neurons, and ameliorates their morphology, neuronal activity and synaptic ultrastructure. Moreover, we provide evidence for attenuation of motor and exploratory impairment in Mecp2 knock-out mice upon trehalose administration. Overall, our findings open new perspectives for neurodevelopmental disorders therapies based on the concept of autophagy modulation.
Keywords: Autophagy; MeCP2; Metabolism; Neurons; Rett Syndrome.
© 2024. The Author(s).
Conflict of interest statement
Figures






References
-
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY (1999) Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23:185–188 - PubMed
-
- Bedogni F, Cobolli Gigli C, Pozzi D, Rossi RL, Scaramuzza L, Rossetti G, Pagani M, Kilstrup-Nielsen C, Matteoli M, Landsberger N (2016) Defects during mecp2 null embryonic cortex development precede the onset of overt neurological symptoms. Cereb Cortex 26:2517–2529 - PubMed
-
- Bingol B (2018) Autophagy and lysosomal pathways in nervous system disorders. Mol Cell Neurosci 91:167–208 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

