SOX4 facilitates brown fat development and maintenance through EBF2-mediated thermogenic gene program in mice
- PMID: 39402212
- PMCID: PMC11893884
- DOI: 10.1038/s41418-024-01397-0
SOX4 facilitates brown fat development and maintenance through EBF2-mediated thermogenic gene program in mice
Abstract
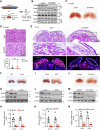
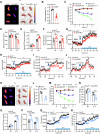
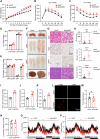
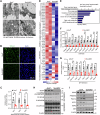
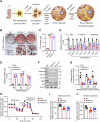
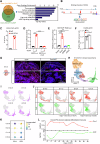
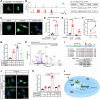
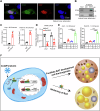
Brown adipose tissue (BAT) is critical for non-shivering thermogenesis making it a promising therapeutic strategy to combat obesity and metabolic disease. However, the regulatory mechanisms underlying brown fat formation remain incompletely understood. Here, we found SOX4 is required for BAT development and thermogenic program. Depletion of SOX4 in BAT progenitors (Sox4-MKO) or brown adipocytes (Sox4-BKO) resulted in whitened BAT and hypothermia upon acute cold exposure. The reduced thermogenic capacity of Sox4-MKO mice increases their susceptibility to diet-induced obesity. Conversely, overexpression of SOX4 in BAT enhances thermogenesis counteracting diet-induced obesity. Mechanistically, SOX4 activates the transcription of EBF2, which determines brown fat fate. Moreover, phosphorylation of SOX4 at S235 by PKA facilitates its nuclear translocation and EBF2 transcription. Further, SOX4 cooperates with EBF2 to activate transcriptional programs governing thermogenic gene expression. These results demonstrate that SOX4 serves as an upstream regulator of EBF2, providing valuable insights into BAT development and thermogenic function maintenance.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics: All mouse experiments were approved and complied with the guidelines of the Institutional Animal Care and Research Advisory Committee at Xiamen University, China (Permit Number: XMULAC20230105).
Figures
References
MeSH terms
Substances
Grants and funding
- 82170266/National Natural Science Foundation of China (National Science Foundation of China)
- 82272944/National Natural Science Foundation of China (National Science Foundation of China)
- 32371223/National Natural Science Foundation of China (National Science Foundation of China)
- 32071150/National Natural Science Foundation of China (National Science Foundation of China)
- 3502Z20224021/Xiamen Municipal Bureau of Science and Technology (Xiamen Science and Technology Bureau)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

