Brown fat ATP-citrate lyase links carbohydrate availability to thermogenesis and guards against metabolic stress
- PMID: 39402290
- PMCID: PMC11841677
- DOI: 10.1038/s42255-024-01143-3
Brown fat ATP-citrate lyase links carbohydrate availability to thermogenesis and guards against metabolic stress
Abstract
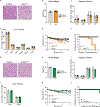
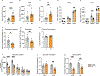
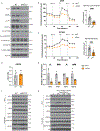
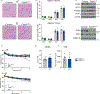
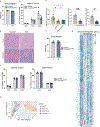
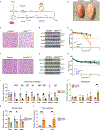
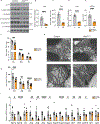
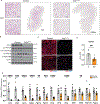
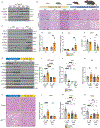
Brown adipose tissue (BAT) engages futile fatty acid synthesis-oxidation cycling, the purpose of which has remained elusive. Here, we show that ATP-citrate lyase (ACLY), which generates acetyl-CoA for fatty acid synthesis, promotes thermogenesis by mitigating metabolic stress. Without ACLY, BAT overloads the tricarboxylic acid cycle, activates the integrated stress response (ISR) and suppresses thermogenesis. ACLY's role in preventing BAT stress becomes critical when mice are weaned onto a carbohydrate-plentiful diet, while removing dietary carbohydrates prevents stress induction in ACLY-deficient BAT. ACLY loss also upregulates fatty acid synthase (Fasn); yet while ISR activation is not caused by impaired fatty acid synthesis per se, deleting Fasn and Acly unlocks an alternative metabolic programme that overcomes tricarboxylic acid cycle overload, prevents ISR activation and rescues thermogenesis. Overall, we uncover a previously unappreciated role for ACLY in mitigating mitochondrial stress that links dietary carbohydrates to uncoupling protein 1-dependent thermogenesis and provides fundamental insight into the fatty acid synthesis-oxidation paradox in BAT.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


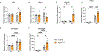






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

