CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody
- PMID: 39402718
- PMCID: PMC11485749
- DOI: 10.1080/19420862.2024.2410316
CD200R1 immune checkpoint blockade by the first-in-human anti-CD200R1 antibody 23ME-00610: molecular mechanism and engineering of a surrogate antibody
Abstract
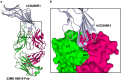
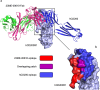
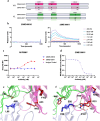
Human CD200R1 (hCD200R1), an immune inhibitory receptor expressed predominantly on T cells and myeloid cells, was identified as a promising immuno-oncology target by the 23andMe database. Blockade of CD200R1-dependent signaling enhances T cell-mediated antitumor activity in vitro and in vivo. 23ME-00610 is a potential first-in-class, humanized IgG1 investigational antibody that binds hCD200R1 with high affinity. We have previously shown that 23ME-00610 inhibits the hCD200R1 immune checkpoint function. Herein, we dissect the molecular mechanism of 23ME-00610 blockade of hCD200R1 by solving the crystal structure of 23ME-00610 Fab in complex with hCD200R1 and performing mutational studies, which show 23ME-00610 blocks the interaction between hCD200 and hCD200R1 through steric hindrance. However, 23ME-00610 does not bind CD200R1 of preclinical species such as cynomolgus monkey MfCD200R1. To enable preclinical toxicology studies of CD200R1 blockade in a pharmacologically relevant non-clinical species, we engineered a surrogate antibody with high affinity toward MfCD200R1. We used phage display libraries of 23ME-00610 variants with individual CDR residues randomized to all 20 amino acids, from which we identified mutations that switched on MfCD200R1 binding. Structural analysis suggests how the surrogate, named 23ME-00611, acquires the ortholog binding ability at the equivalent epitope of 23ME-00610. This engineering approach does not require a priori knowledge of structural and functional mapping of antibody-antigen interaction and thus is generally applicable for therapeutic antibody development when desired ortholog binding is lacking. These findings provide foundational insights as 23ME-00610 advances in clinical studies to gain understanding of the hCD200R1 immune checkpoint as a target in immuno-oncology.
Keywords: 23ME-00610; CD200; CD200R1; cancer immunotherapy; deep mutational scan; immune checkpoint.
Conflict of interest statement
AD, LL, GF, YH: Employees of 23andMe. OG: Employees of Proteros Biostructures. BC, CM, AJH, and SJB were employees of 23andMe at the time this work was performed.
Figures



References
-
- Wright GJ, Cherwinski H, Foster-Cuevas M, Brooke G, Puklavec MJ, Bigler M, Song Y, Jenmalm M, Gorman D, McClanahan T, et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J Immunol. 2003;171(6):3034–12. doi: 10.4049/jimmunol.171.6.3034. - DOI - PubMed
-
- McWhirter JR, Kretz-Rommel A, Saven A, Maruyama T, Potter KN, Mockridge CI, Ravey EP, Qin F, Bowdish KS. Antibodies selected from combinatorial libraries block a tumor antigen that plays a key role in immunomodulation. Proc Natl Acad Sci USA. 2006;103(4):1041–1046. doi: 10.1073/pnas.0510081103. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
