The USP1 Inhibitor KSQ-4279 Overcomes PARP Inhibitor Resistance in Homologous Recombination-Deficient Tumors
- PMID: 39402989
- PMCID: PMC11474170
- DOI: 10.1158/0008-5472.CAN-24-0293
The USP1 Inhibitor KSQ-4279 Overcomes PARP Inhibitor Resistance in Homologous Recombination-Deficient Tumors
Abstract
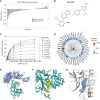
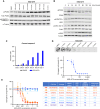
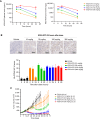
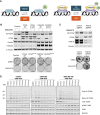
Defects in DNA repair pathways play a pivotal role in tumor evolution and resistance to therapy. At the same time, they create vulnerabilities that render tumors dependent on the remaining DNA repair processes. This phenomenon is exemplified by the clinical activity of PARP inhibitors in tumors with homologous recombination (HR) repair defects, such as tumors with inactivating mutations in BRCA1 or BRCA2. However, the development of resistance to PARP inhibitors in BRCA-mutant tumors represents a high unmet clinical need. In this study, we identified deubiquitinase ubiquitin-specific peptidase-1 (USP1) as a critical dependency in tumors with BRCA mutations or other forms of HR deficiency and developed KSQ-4279, the first potent and selective USP1 inhibitor to enter clinical testing. The combination of KSQ-4279 with a PARP inhibitor was well tolerated and induced durable tumor regression across several patient-derived PARP-resistant models. These findings indicate that USP1 inhibitors represent a promising therapeutic strategy for overcoming PARP inhibitor resistance in patients with BRCA-mutant/HR-deficient tumors and support continued testing in clinical trials. Significance: KSQ-4279 is a potent and selective inhibitor of USP1 that induces regression of PARP inhibitor-resistant tumors when dosed in combination with PARP inhibitors, addressing an unmet clinical need for BRCA-mutant tumors.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
E. Krall reports personal fees from Merck Sharp and Dohme LLC outside the submitted work, as well as a patent 11485736 issued. E. Chipumuro reports other support from KSQ Therapeutics outside the submitted work. K. Sinkevicius reports employment with KSQ Therapeutics and receiving KSQ Therapeutics stock options. P.C. Gokhale reports grants from KSQ Therapeutics, Inc. during the conduct of the study, as well as grants from Kymera Therapeutics, Pfizer Inc., Treeline Biosciences, and Arvinas outside the submitted work. U.A. Matulonis reports personal fees from Tango Therapeutics, ProfoundBio, NextCure, Eisai, ImmunoGen, AbbVie, Alkermes, Novartis, and GSK outside the submitted work. J.F. Liu reports personal fees from AstraZeneca, Bristol Myers Squibb, Clovis Oncology, Daiichi Sankyo, Eisai, Genentech/Roche, GlaxoSmithKline, Regeneron Therapeutics, and Zentalis Pharmaceuticals outside the submitted work. M. Schlabach reports a patent for US 2021/0115049 A1 pending. F. Stegmeier reports full-time employment with KSQ Therapeutics during the time of this work, as well as being on the Board of Directors and ownership of stocks in KSQ Therapeutics. A.A. Wylie reports personal fees from KSQ Therapeutics outside the submitted work, as well as a patent for WO2021163530A1 issued, WO20230203046A1 issued, and WO2023147311A1 issued. J. Brenneman, J.J. Ali, S. Shenker, M. McGuire, P. Grasberger, Y. Mishina, E. Krall, J. Hixon, E. Chipumuro, K. Sinkevicius, A. Olaharski, H. Liu, J. Wilt, M. Schlabach, and F. Stegmeier conducted this research while employed by KSQ Therapeutics. No disclosures were reported by the other authors.
Figures

References
-
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability-an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220–8. - PubMed
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. - PubMed
-
- Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006;25:5864–74. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

