Bayesian Variable Shrinkage and Selection in Compositional Data Regression: Application to Oral Microbiome
- PMID: 39403125
- PMCID: PMC11470902
- DOI: 10.1007/s41096-024-00194-9
Bayesian Variable Shrinkage and Selection in Compositional Data Regression: Application to Oral Microbiome
Abstract
Microbiome studies generate multivariate compositional responses, such as taxa counts, which are strictly non-negative, bounded, residing within a simplex, and subject to unit-sum constraint. In presence of covariates (which can be moderate to high dimensional), they are popularly modeled via the Dirichlet-Multinomial (D-M) regression framework. In this paper, we consider a Bayesian approach for estimation and inference under a D-M compositional framework, and present a comparative evaluation of some state-of-the-art continuous shrinkage priors for efficient variable selection to identify the most significant associations between available covariates, and taxonomic abundance. Specifically, we compare the performances of the horseshoe and horseshoe+ priors (with the benchmark Bayesian lasso), utilizing Hamiltonian Monte Carlo techniques for posterior sampling, and generating posterior credible intervals. Our simulation studies using synthetic data demonstrate excellent recovery and estimation accuracy of sparse parameter regime by the continuous shrinkage priors. We further illustrate our method via application to a motivating oral microbiome data generated from the NYC-Hanes study. RStan implementation of our method is made available at the GitHub link: (https://github.com/dattahub/compshrink).
Keywords: Bayesian; Compositional data; Dirichlet; Generalized Dirichlet; Horseshoe; Large p; Shrinkage prior; Sparse probability vectors; Stick-breaking.
© The Author(s) 2024.
Conflict of interest statement
Conflict of interestThe authors declare that they have no conflict of interest.
Figures

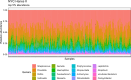
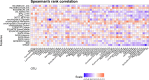
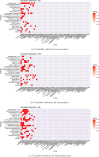
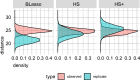
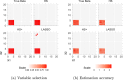
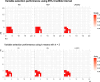
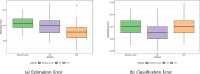
References
-
- Betancourt M, Byrne S, Livingstone S, Girolami M (2017) The geometric foundations of Hamiltonian Monte Carlo. Bernoulli 23(4A):2257–2298. 10.3150/16-BEJ810 - DOI
-
- Bhadra A, Datta J, Polson NG, Willard B (2016) Default bayesian analysis with global-local shrinkage priors. Biometrika 103(4):955–969
LinkOut - more resources
Full Text Sources
