Magnetic nanobead assisted the dual targets driven fluorescent biosensor based on SPEXPAR and MNAzyme for the olfactory marker protein detection
- PMID: 39403314
- PMCID: PMC11471671
- DOI: 10.1016/j.mtbio.2024.101272
Magnetic nanobead assisted the dual targets driven fluorescent biosensor based on SPEXPAR and MNAzyme for the olfactory marker protein detection
Abstract
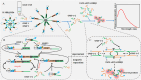
Fig. 1: Schematic illustration of the principle of the Dual targets driven SPEXPAR assisted MNAzyme (D-S-M) fluorescent biosensor for olfactory marker protein (OMP) detection. A. Workflow for detection of OMP in nasal swab. B. Isothermal Self-Primer EXPonential Amplification Reaction (SPEXPAR) amplification. C. The production of fluorescent signal by Multicomponent Nuclear Acid Enzyme (MNAzyme). The signal of OMP was amplified and changed into the detectable fluorescence signal based on the reactions of SPEXPAR and MNAzyme in the D-S-M fluorescence biosensor. The qualitative or quantitative analysis of OMP can be measured by the analysis of the fluorescence intensity.Image 1.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Whitcroft K.L., Alobid I., Altundag A., Andrews P., Carrie S., Fahmy M., Fjaeldstad A.W., Gane S., Hopkins C., Hsieh J.W., Huart C., Hummel T., Konstantinidis I., Landis B.N., Mori E., Mullol J., Philpott C., Poulios A., Vodička J., Ward V.M. International clinical assessment of smell: an international, cross-sectional survey of current practice in the assessment of olfaction. Clin. Otolaryngol. 2024;49:220–234. doi: 10.1111/coa.14123. - DOI - PubMed
LinkOut - more resources
Full Text Sources

