Assembly of Mitochondrial Genomes Using Nanopore Long-Read Technology in Three Sea Chubs (Teleostei: Kyphosidae)
- PMID: 39403800
- PMCID: PMC11646298
- DOI: 10.1111/1755-0998.14034
Assembly of Mitochondrial Genomes Using Nanopore Long-Read Technology in Three Sea Chubs (Teleostei: Kyphosidae)
Abstract
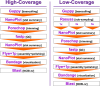
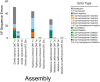
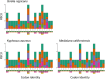
Complete mitochondrial genomes have become markers of choice to explore phylogenetic relationships at multiple taxonomic levels and they are often assembled using whole genome short-read sequencing. Herein, using three species of sea chubs as an example, we explored the accuracy of mitochondrial chromosomes assembled using Oxford Nanopore Technology (ONT) Kit 14 R10.4.1 long reads at different sequencing depths (high, low and very low or genome skimming) by comparing them to 'gold' standard reference mitochondrial genomes assembled using Illumina NovaSeq short reads. In two species of sea chubs, Girella nigricans and Kyphosus azureus, ONT long-read assembled mitochondrial genomes at high sequencing depths (> 25× whole [nuclear] genome) were identical to their respective short-read assembled mitochondrial genomes. Not a single 'homopolymer insertion', 'homopolymer deletion', 'simple substitution', 'single insertion', 'short insertion', 'single deletion' or 'short deletion' were detected in the long-read assembled mitochondrial genomes after aligning each one of them to their short-read counterparts. In turn, in a third species, Medialuna californiensis, a 25× sequencing depth long-read assembled mitochondrial genome was 14 nucleotides longer than its short-read counterpart. The difference in total length between the latter two assemblies was due to the presence of a short motif 14 bp long that was repeated (twice) in the long read but not in the short-read assembly. Read subsampling at a sequencing depth of 1× resulted in the assembly of partial or complete mitochondrial genomes with numerous errors, including, among others, simple indels, and indels at homopolymer regions. At 3× and 5× subsampling, genomes were identical (perfect) or almost identical (quasiperfect, 99.5% over 16,500 bp) to their respective Illumina assemblies. The newly assembled mitochondrial genomes exhibit identical gene composition and organisation compared with cofamilial species and a phylomitogenomic analysis based on translated protein-coding genes suggested that the family Kyphosidae is not monophyletic. The same analysis detected possible cases of misidentification of mitochondrial genomes deposited in GenBank. This study demonstrates that perfect (complete and fully accurate) or quasiperfect (complete but with a single or a very few errors) mitochondrial genomes can be assembled at high (> 25×) and low (3-5×) but not very low (1×, genome skimming) sequencing depths using ONT long reads and the latest ONT chemistries (Kit 14 and R10.4.1 flowcells with SUP basecalling). The newly assembled and annotated mitochondrial genomes can be used as a reference in environmental DNA studies focusing on bioprospecting and biomonitoring of these and other coastal species experiencing environmental insult. Given the small size of the sequencing device and low cost, we argue that ONT technology has the potential to improve access to high-throughput sequencing technologies in low- and moderate-income countries.
Keywords: long‐read sequencing; nanopore; rudderfish.
© 2024 The Author(s). Molecular Ecology Resources published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Ballard, J. W. O. , and Rand D. M.. 2005. “The Population Biology of Mitochondrial DNA and Its Phylogenetic Implications.” Annual Review of Ecology, Evolution, and Systematics 36: 621–642.
MeSH terms
LinkOut - more resources
Full Text Sources

