DeepPhylo: Phylogeny-Aware Microbial Embeddings Enhanced Predictive Accuracy in Human Microbiome Data Analysis
- PMID: 39403892
- PMCID: PMC11615782
- DOI: 10.1002/advs.202404277
DeepPhylo: Phylogeny-Aware Microbial Embeddings Enhanced Predictive Accuracy in Human Microbiome Data Analysis
Abstract
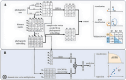
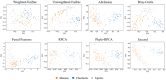
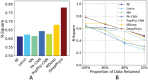
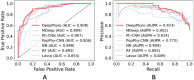
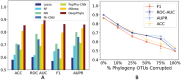
Microbial data analysis poses significant challenges due to its high dimensionality, sparsity, and compositionality. Recent advances have shown that integrating abundance and phylogenetic information is an effective strategy for uncovering robust patterns and enhancing the predictive performance in microbiome studies. However, existing methods primarily focus on the hierarchical structure of phylogenetic trees, overlooking the evolutionary distances embedded within them. This study introduces DeepPhylo, a novel method that employs phylogeny-aware amplicon embeddings to effectively integrate abundance and phylogenetic information. DeepPhylo improves both the unsupervised discriminatory power and supervised predictive accuracy of microbiome data analysis. Compared to the existing methods, DeepPhylo demonstrates superiority in informing biologically relevant insights across five real-world microbiome use cases, including clustering of skin microbiomes, prediction of host chronological age and gender, diagnosis of inflammatory bowel disease (IBD) across 15 studies, and multilabel disease classification.
Keywords: beta‐diversity; deep learning; microbiome; phylogeny.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
