A High-Throughput Drug Repurposing Strategy to Treat TBX2 and/or TBX3 Dependent Cancers
- PMID: 39403898
- PMCID: PMC11474296
- DOI: 10.1002/cam4.70303
A High-Throughput Drug Repurposing Strategy to Treat TBX2 and/or TBX3 Dependent Cancers
Abstract
Background: The highly homologous T-box transcription factors TBX2 and TBX3 are critical for embryonic development, and their overexpression in postnatal tissues contributes to a wide range of malignancies, including melanoma and rhabdomyosarcoma. Importantly, when TBX2 and TBX3 are depleted in cancers where they are overexpressed, the malignant phenotype is inhibited, and they have therefore been regarded as druggable targets. However, the time and costs associated with de novo drug development are challenging and result in drugs that are costly, especially for patients in low- and middle-income countries. In the current study, we therefore combined a targeted and drug repurposing approach to identify drugs that are expected to be more efficacious and cost-effective with significantly reduced side effects.
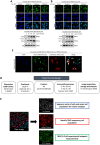
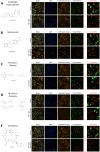
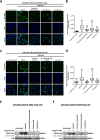
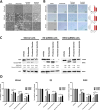
Methods: A high-throughput cell-based immunofluorescence screen was performed to identify drugs in the Pharmakon 1600 drug library that can negatively regulate TBX2 and/or TBX3 levels. "Hit" drugs were validated for their effect on TBX2/TBX3 levels and cytotoxicity in TBX2/TBX3-dependent melanoma and rhabdomyosarcoma cells. To this end, immunofluorescence, western blotting, quantitative real-time PCR, and MTT cell viability assays were performed.
Results: Niclosamide, piroctone olamine, and pyrvinium pamoate, were identified as TBX2 and/or TBX3-targeting drugs, and they exhibited cytotoxicity in a TBX2/TBX3-dependent manner. Furthermore, these "Hit" drugs were shown to induce senescence and/or apoptosis.
Conclusions: Niclosamide, piroctone olamine, and pyrvinium pamoate are promising, cost-effective therapeutic agents for the treatment of TBX2/TBX3-dependent cancers.
Keywords: T‐box factors; drug repurposing; high‐throughput screen; transcription factors.
© 2024 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
- Newton RCUK PhD Partnering Scheme
- South African Medical Research Council, Self-Initiated Research Grant
- National Research Foundation of South Africa, Competitive Programme for Rated Researchers
- International Centre for Genetic Engineering and Biotechnology, Collaborative Research Programme
- University of Cape Town, UCT Vision 2030 Grand Challenges Programme
LinkOut - more resources
Full Text Sources

