Autoinflammation in patients with leukocytic CBL loss of heterozygosity is caused by constitutive ERK-mediated monocyte activation
- PMID: 39403923
- PMCID: PMC11475086
- DOI: 10.1172/JCI181604
Autoinflammation in patients with leukocytic CBL loss of heterozygosity is caused by constitutive ERK-mediated monocyte activation
Abstract
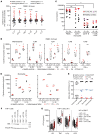
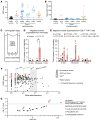
Patients heterozygous for germline CBL loss-of-function (LOF) variants can develop myeloid malignancy, autoinflammation, or both, if some or all of their leukocytes become homozygous for these variants through somatic loss of heterozygosity (LOH) via uniparental isodisomy. We observed an upregulation of the inflammatory gene expression signature in whole blood from these patients, mimicking monogenic inborn errors underlying autoinflammation. Remarkably, these patients had constitutively activated monocytes that secreted 10 to 100 times more inflammatory cytokines than those of healthy individuals and CBL LOF heterozygotes without LOH. CBL-LOH hematopoietic stem and progenitor cells (HSPCs) outgrew the other cells, accounting for the persistence of peripheral monocytes homozygous for the CBL LOF variant. ERK pathway activation was required for the excessive production of cytokines by both resting and stimulated CBL-LOF monocytes, as shown in monocytic cell lines. Finally, we found that about 1 in 10,000 individuals in the UK Biobank were heterozygous for CBL LOF variants and that these carriers were at high risk of hematological and inflammatory conditions.
Keywords: Autoimmunity; Immunology; Innate immunity; Monocytes.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

