Surface-engineered bacteria in drug development
- PMID: 39403960
- PMCID: PMC11474283
- DOI: 10.1111/1751-7915.70033
Surface-engineered bacteria in drug development
Abstract
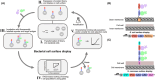
Bacterial surface display in combination with fluorescence-activated cell sorting is a versatile and robust system and an interesting alternative approach to phage display for the generation of therapeutic affinity proteins. The system enables real-time monitoring and sorting of cell populations, which presents unique possibilities for drug development. It has been used to develop several affibody molecules currently being evaluated preclinically for the treatment and diagnosis of, for example, cancer and neurodegenerative diseases. Additionally, it can be implemented in other areas of drug design, such as for mapping epitopes and evolving enzyme specificities.
© 2024 The Author(s). Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
S. S. is a minority shareholder in Affibody AB. C. D. L. and J. L. declare no conflict of interest.
Figures

References
-
- Altai, M. , Strand, J. , Rosik, D. , Selvaraju, R.K. , Eriksson Karlström, A. , Orlova, A. et al. (2013) Influence of nuclides and chelators on imaging using affibody molecules: comparative evaluation of recombinant affibody molecules site‐specifically labeled with 68Ga and 111In via maleimido derivatives of DOTA and NODAGA. Bioconjugate Chemistry, 24, 1102–1109. - PubMed
-
- Andersson, K.G. , Persson, J. , Ståhl, S. & Löfblom, J. (2019) Autotransporter‐mediated display of a naïve affibody library on the outer membrane of Escherichia coli . Biotechnology Journal, 14, 1–8. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

