Chromatin Accessibility and Gene Expression Vary Between a New and Evolved Autopolyploid of Arabidopsis arenosa
- PMID: 39404085
- PMCID: PMC11518924
- DOI: 10.1093/molbev/msae213
Chromatin Accessibility and Gene Expression Vary Between a New and Evolved Autopolyploid of Arabidopsis arenosa
Abstract
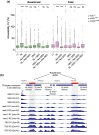
Polyploids arise from whole-genome duplication (WGD) events, which have played important roles in genome evolution across eukaryotes. WGD can increase genome complexity, yield phenotypic novelty, and influence adaptation. Neo-polyploids have been reported to often show seemingly stochastic epigenetic and transcriptional changes, but this leaves open the question whether these changes persist in evolved polyploids. A powerful approach to address this is to compare diploids, neo-polyploids, and evolved polyploids of the same species. Arabidopsis arenosa is a species that allows us to do this-natural diploid and autotetraploid populations exist, while neo-tetraploids can be artificially generated. Here, we use ATAC-seq to assay local chromatin accessibility, and RNA-seq to study gene expression on matched leaf and petal samples from diploid, neo-tetraploid and evolved tetraploid A. arenosa. We found over 8,000 differentially accessible chromatin regions across all samples. These are largely tissue specific and show distinct trends across cytotypes, with roughly 70% arising upon WGD. Interestingly, only a small proportion is associated with expression changes in nearby genes. However, accessibility variation across cytotypes associates strongly with the number of nearby transposable elements. Relatively few genes were differentially expressed upon genome duplication, and ∼60% of these reverted to near-diploid levels in the evolved tetraploid, suggesting that most initial perturbations do not last. Our results provide new insights into how epigenomic and transcriptional mechanisms jointly respond to genome duplication and subsequent evolution of autopolyploids, and importantly, show that one cannot be directly predicted from the other.
Keywords: Arabidopsis arenosa; chromatin; epigenome; gene expression; polyploids.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of interest The authors declare that they have no competing interests.
Figures




References
-
- Alexandre CM, Urton JR, Jean-Baptiste K, Huddleston J, Dorrity MW, Cuperus JT, Sullivan AM, Bemm F, Jolic D, Arsovski AA, et al. Complex relationships between chromatin accessibility, sequence divergence, and gene expression in Arabidopsis thaliana. Mol Biol Evol. 2018:35(4):837–854. 10.1093/molbev/msx326. - DOI - PMC - PubMed
-
- Allario T, Brumos J, Colmenero-Flores JM, Tadeo F, Froelicher Y, Talon M, Navarro L, Ollitrault P, Morillon R. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J Exp Bot. 2011:62(8):2507–2519. 10.1093/jxb/erq467. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

