Sentinel Surveillance reveals phylogenetic diversity and detection of linear plasmids harboring vanA and optrA among enterococci collected in the United States
- PMID: 39404260
- PMCID: PMC11539240
- DOI: 10.1128/aac.00591-24
Sentinel Surveillance reveals phylogenetic diversity and detection of linear plasmids harboring vanA and optrA among enterococci collected in the United States
Abstract
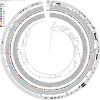
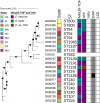
Enterococcus faecalis and Enterococcus faecium are frequent causes of healthcare-associated infections. Antimicrobial-resistant enterococci pose a serious public health threat, particularly vancomycin-resistant enterococci (VRE), for which treatment options are limited. The Centers for Disease Control and Prevention's Division of Healthcare Quality Promotion Sentinel Surveillance system conducted surveillance from 2018 to 2019 to evaluate antimicrobial susceptibility profiles and molecular epidemiology of 205 E. faecalis and 180 E. faecium clinical isolates collected from nine geographically diverse sites in the United States. Whole genome sequencing revealed diverse genetic lineages, with no single sequence type accounting for more than 15% of E. faecalis or E. faecium. Phylogenetic analysis distinguished E. faecium from 19 E. lactis (previously known as E. faecium clade B). Resistance to vancomycin was 78.3% among E. faecium, 7.8% among E. faecalis, and did not occur among E. lactis isolates. Resistance to daptomycin and linezolid was rare: E. faecium (5.6%, 0.6%, respectively), E. faecalis (2%, 2%), and E. lactis (5.3%, 0%). All VRE harbored the vanA gene. Three of the seven isolates that were not susceptible to linezolid harbored optrA, one chromosomally located and two on linear plasmids that shared a conserved backbone with other multidrug-resistant conjugative linear plasmids. One of these isolates contained optrA and vanA co-localized on the linear plasmid. By screening all enterococci, 20% of E. faecium were predicted to harbor linear plasmids, whereas none were predicted among E. faecalis or E. lactis. Continued surveillance is needed to assess the future emergence and spread of antimicrobial resistance by linear plasmids and other mechanisms.IMPORTANCEThis work confirms prior reports of E. faecium showing higher levels of resistance to more antibiotics than E. faecalis and identifies that diverse sequence types are contributing to enterococcal infections in the United States. All VRE harbored the vanA gene. We present the first report of the linezolid resistance gene optrA on linear plasmids in the United States, one of which co-carried a vanA cassette. Additional studies integrating epidemiological, antimicrobial susceptibility, and genomic methods to characterize mechanisms of resistance, including the role of linear plasmids, will be critical to understanding the changing landscape of enterococci in the United States.
Keywords: Enterococcus faecalis; Enterococcus faecium; daptomycin; linear plasmid; linezolid; optrA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- CDC . 2019. Antibiotic resistance threats in the United States, 2019. Atlanta, GA: Department of Health and Human Services C, Department of Health and Human Services, CDC
-
- Jernigan JA, Hatfield KM, Wolford H, Nelson RE, Olubajo B, Reddy SC, McCarthy N, Paul P, McDonald LC, Kallen A, Fiore A, Craig M, Baggs J. 2020. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012-2017. N Engl J Med 382:1309–1319. doi: 10.1056/NEJMoa1914433 - DOI - PMC - PubMed
-
- CDC . 2022. COVID-19: U.S. impact on antimicrobial resistance, special report 2022. doi: 10.15620/cdc:117915 - DOI
-
- Gargis AS, Spicer LM, Kent AG, Zhu W, Campbell D, McAllister G, Ewing TO, Albrecht V, Stevens VA, Sheth M, Padilla J, Batra D, Johnson JK, Halpin AL, Rasheed JK, Elkins CA, Karlsson M, Lutgring JD. 2021. Sentinel surveillance reveals emerging daptomycin-resistant ST736 Enterococcus faecium and multiple mechanisms of linezolid resistance in enterococci in the United States. Front Microbiol 12:807398. doi: 10.3389/fmicb.2021.807398 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

