The structural and mechanistic bases for the viral resistance to allosteric HIV-1 integrase inhibitor pirmitegravir
- PMID: 39404354
- PMCID: PMC11559089
- DOI: 10.1128/mbio.00465-24
The structural and mechanistic bases for the viral resistance to allosteric HIV-1 integrase inhibitor pirmitegravir
Erratum in
-
Erratum for Dinh et al., "The structural and mechanistic bases for the viral resistance to allosteric HIV-1 integrase inhibitor pirmitegravir".mBio. 2025 Dec 10;16(12):e0310625. doi: 10.1128/mbio.03106-25. Epub 2025 Nov 4. mBio. 2025. PMID: 41186432 Free PMC article. No abstract available.
Abstract
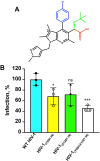
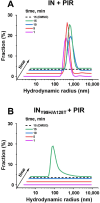
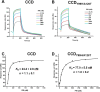
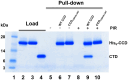
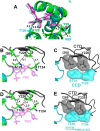
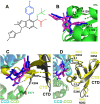
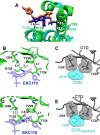
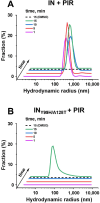
Allosteric HIV-1 integrase (IN) inhibitors (ALLINIs) are investigational antiretroviral agents that potently impair virion maturation by inducing hyper-multimerization of IN and inhibiting its interaction with viral genomic RNA. The pyrrolopyridine-based ALLINI pirmitegravir (PIR) has recently advanced into phase 2a clinical trials. Previous cell culture-based viral breakthrough assays identified the HIV-1(Y99H/A128T IN) variant that confers substantial resistance to this inhibitor. Here, we have elucidated the unexpected mechanism of viral resistance to PIR. Although both Tyr99 and Ala128 are positioned within the inhibitor binding V-shaped cavity at the IN catalytic core domain (CCD) dimer interface, the Y99H/A128T IN mutations did not substantially affect the direct binding of PIR to the CCD dimer or functional oligomerization of full-length IN. Instead, the drug-resistant mutations introduced a steric hindrance at the inhibitor-mediated interface between CCD and C-terminal domain (CTD) and compromised CTD binding to the CCDY99H/A128T + PIR complex. Consequently, full-length INY99H/A128T was substantially less susceptible to the PIR-induced hyper-multimerization than the WT protein, and HIV-1(Y99H/A128T IN) conferred >150-fold resistance to the inhibitor compared with the WT virus. By rationally modifying PIR, we have developed its analog EKC110, which readily induced hyper-multimerization of INY99H/A128T in vitro and was ~14-fold more potent against HIV-1(Y99H/A128T IN) than the parent inhibitor. These findings suggest a path for developing improved PIR chemotypes with a higher barrier to resistance for their potential clinical use.IMPORTANCEAntiretroviral therapies save the lives of millions of people living with HIV (PLWH). However, the evolution of multi-drug-resistant viral phenotypes is a major clinical problem, and there are limited or no treatment options for heavily treatment-experienced PLWH. Allosteric HIV-1 integrase inhibitors (ALLINIs) are a novel class of antiretroviral compounds that work by a unique mechanism of binding to the non-catalytic site on the viral protein and inducing aberrant integrase multimerization. Accordingly, ALLINIs potently inhibit both wild-type HIV-1 and all drug-resistant viral phenotypes that have so far emerged against currently used therapies. Pirmitegravir, a highly potent and safe investigational ALLINI, is currently advancing through clinical trials. Here, we have elucidated the structural and mechanistic bases behind the emergence of HIV-1 integrase mutations in infected cells that confer resistance to pirmitegravir. In turn, our findings allowed us to rationally develop an improved ALLINI with substantially enhanced potency against the pirmitegravir-resistant virus.
Keywords: ALLINI; HIV-1 integrase; antiretroviral drug; pirmitegravir.
Conflict of interest statement
Kyungjin Kim was the chief executive officer of ST Pharm Co. Ltd. No other authors declare a potential conflict of interest.
Figures

Update of
-
The structural and mechanistic bases for the viral resistance to allosteric HIV-1 integrase inhibitor pirmitegravir.bioRxiv [Preprint]. 2024 Jan 26:2024.01.26.577387. doi: 10.1101/2024.01.26.577387. bioRxiv. 2024. Update in: mBio. 2024 Nov 13;15(11):e0046524. doi: 10.1128/mbio.00465-24. PMID: 38328097 Free PMC article. Updated. Preprint.
References
-
- Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, Marchand D, Bardiot D, Van der Veken NJ, Van Remoortel B, Strelkov SV, De Maeyer M, Chaltin P, Debyser Z. 2010. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat Chem Biol 6:442–448. doi: 10.1038/nchembio.370 - DOI - PubMed
-
- Patel D, Antwi J, Koneru PC, Serrao E, Forli S, Kessl JJ, Feng L, Deng N, Levy RM, Fuchs JR, Olson AJ, Engelman AN, Bauman JD, Kvaratskhelia M, Arnold E. 2016. A new class of allosteric HIV-1 integrase inhibitors identified by crystallographic fragment screening of the catalytic core domain. J Biol Chem 291:23569–23577. doi: 10.1074/jbc.M116.753384 - DOI - PMC - PubMed
-
- Maehigashi T, Ahn S, Kim UI, Lindenberger J, Oo A, Koneru PC, Mahboubi B, Engelman AN, Kvaratskhelia M, Kim K, Kim B. 2021. A highly potent and safe pyrrolopyridine-based allosteric HIV-1 integrase inhibitor targeting host LEDGF/p75-integrase interaction site. PLoS Pathog 17:e1009671. doi: 10.1371/journal.ppat.1009671 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
