Human Umbilical Cord-Mesenchymal Stem Cells Promote Extracellular Matrix Remodeling in Microglia
- PMID: 39404427
- PMCID: PMC11475221
- DOI: 10.3390/cells13191665
Human Umbilical Cord-Mesenchymal Stem Cells Promote Extracellular Matrix Remodeling in Microglia
Abstract
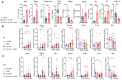
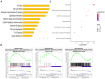
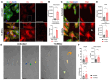
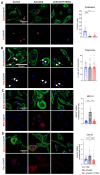
Human mesenchymal stem cells modulate the immune response and are good candidates for cell therapy in neuroinflammatory brain disorders affecting both adult and premature infants. Recent evidence indicates that through their secretome, mesenchymal stem cells direct microglia, brain-resident immune cells, toward pro-regenerative functions, but the mechanisms underlying microglial phenotypic transition are still under investigation. Using an in vitro coculture approach combined with transcriptomic analysis, we identified the extracellular matrix as the most relevant pathway altered by the human mesenchymal stem cell secretome in the response of microglia to inflammatory cytokines. We confirmed extracellular matrix remodeling in microglia exposed to the mesenchymal stem cell secretome via immunofluorescence analysis of the matrix component fibronectin and the extracellular crosslinking enzyme transglutaminase-2. Furthermore, an analysis of hallmark microglial functions revealed that changes in the extracellular matrix enhance ruffle formation by microglia and cell motility. These findings point to extracellular matrix changes, associated plasma membrane remodeling, and enhanced microglial migration as novel mechanisms by which mesenchymal stem cells contribute to the pro-regenerative microglial transition.
Keywords: extracellular matrix; human umbilical cord mesenchymal stem cells; microglia; migration; neuroinflammation.
Conflict of interest statement
Nicola Pelizzi is an employee of Chiesi Farmaceutici S.p.A. All other authors declare that they have no competing interests.
Figures

References
-
- Sobue A., Komine O., Hara Y., Endo F., Mizoguchi H., Watanabe S., Murayama S., Saito T., Saido T.C., Sahara N., et al. Microglial Gene Signature Reveals Loss of Homeostatic Microglia Associated with Neurodegeneration of Alzheimer’s Disease. Acta Neuropathol. Commun. 2021;9:1. doi: 10.1186/s40478-020-01099-x. - DOI - PMC - PubMed
-
- Madry C., Kyrargyri V., Arancibia-Cárcamo I.L., Jolivet R., Kohsaka S., Bryan R.M., Attwell D. Microglial Ramification, Surveillance, and Interleukin-1β Release Are Regulated by the Two-Pore Domain K+ Channel THIK-1. Neuron. 2018;97:299–312.e6. doi: 10.1016/j.neuron.2017.12.002. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

