Insights into human norovirus cultivation in human intestinal enteroids
- PMID: 39404443
- PMCID: PMC11580437
- DOI: 10.1128/msphere.00448-24
Insights into human norovirus cultivation in human intestinal enteroids
Abstract
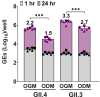
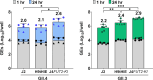
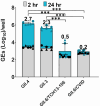
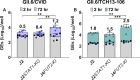
Human noroviruses (HuNoVs) are a significant cause of epidemic and sporadic acute gastroenteritis worldwide. The lack of a reproducible culture system hindered the study of HuNoV replication and pathogenesis for almost a half-century. This barrier was overcome with our successful cultivation of multiple HuNoV strains in human intestinal enteroids (HIEs), which has significantly advanced HuNoV research. We optimized culture media conditions and generated genetically modified HIE cultures to enhance HuNoV replication in HIEs. Building upon these achievements, we now present new insights into this culture system, which involve testing different media, unique HIE lines, and additional virus strains. HuNoV infectivity was evaluated and compared in new HIE models, including HIEs generated from different intestinal segments of individual adult organ donors, HIEs from human intestinal organoids produced from directed differentiation of human embryonic stem cells that were then transplanted and matured in mice before making enteroids (H9tHIEs), genetically engineered (J4FUT2 knock-in [KI], J2STAT1 knockout [KO]) HIEs, as well as HIEs derived from a patient with common variable immunodeficiency (CVID) and from infants. Our findings reveal that small intestinal HIEs, but not colonoids, from adults, H9tHIEs, HIEs from a CVID patient, and HIEs from infants support HuNoV replication with segment and strain-specific differences in viral infection. J4FUT2-KI HIEs exhibit the highest susceptibility to HuNoV infection, allowing the cultivation of a broader range of genogroup I and II HuNoV strains than previously reported. Overall, these results contribute to a deeper understanding of HuNoVs and highlight the transformative potential of HIE cultures in HuNoV research.IMPORTANCEHuman noroviruses (HuNoVs) cause global diarrheal illness and chronic infections in immunocompromised patients. This paper reports approaches for cultivating HuNoVs in secretor positive human intestinal enteroids (HIEs). HuNoV infectivity was compared in new HIE models, including ones from (i) different intestinal segments of single donors, (ii) human embryonic stem cell-derived organoids transplanted into mice, (iii) genetically modified lines, and (iv) a patient with common variable immunodeficiency disease. HIEs from small intestine, but not colon, support HuNoV replication with donor, segment, and strain-specific variations. Unexpectedly, HIEs from one donor are resistant to GII.3 infection. The genetically modified J4FUT2 knock-in (KI) HIEs enable cultivation of a broad range of GI and GII genotypes. New insights into strain-specific differences in HuNoV replication in HIEs support this platform for advancing understanding of HuNoV biology and developing potential therapeutics.
Keywords: HuNoV cultivation; enteroids; organoid growth media; organoids.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Update of
-
Insights into Human Norovirus Cultivation in Human Intestinal Enteroids.bioRxiv [Preprint]. 2024 Sep 19:2024.05.24.595764. doi: 10.1101/2024.05.24.595764. bioRxiv. 2024. Update in: mSphere. 2024 Nov 21;9(11):e0044824. doi: 10.1128/msphere.00448-24. PMID: 38826387 Free PMC article. Updated. Preprint.
References
-
- Pires SM, Fischer-Walker CL, Lanata CF, Devleesschauwer B, Hall AJ, Kirk MD, Duarte ASR, Black RE, Angulo FJ. 2015. Aetiology-specific estimates of the global and regional incidence and mortality of diarrhoeal diseases commonly transmitted through food. PLoS ONE 10:e0142927. doi:10.1371/journal.pone.0142927 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- RP150578, RP170719/American Academy of CPR and First Aid (American Academy of CPR & First Aid, Inc.)
- 1K08DK131326/HHS | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)
- U19 AI116497/AI/NIAID NIH HHS/United States
- P30 DK056338/DK/NIDDK NIH HHS/United States
- P01-AI057788, CA125123/HHS | U.S. Public Health Service (PHS)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

