Combinatorial control of type IVa pili formation by the four polarized regulators MglA, SgmX, FrzS, and SopA
- PMID: 39404445
- PMCID: PMC11580455
- DOI: 10.1128/jb.00108-24
Combinatorial control of type IVa pili formation by the four polarized regulators MglA, SgmX, FrzS, and SopA
Abstract
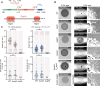
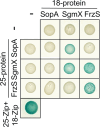
Type IVa pili (T4aP) are widespread and enable bacteria to translocate across surfaces. T4aP engage in cycles of extension, surface adhesion, and retraction, thereby pulling cells forward. Accordingly, the number and localization of T4aP are critical to efficient translocation. Here, we address how T4aP formation is regulated in Myxococcus xanthus, which translocates with a well-defined leading and lagging cell pole using T4aP at the leading pole. This localization is orchestrated by the small GTPase MglA and its downstream effector SgmX that both localize at the leading pole and recruit the PilB extension ATPase to the T4aP machinery at this pole. Here, we identify the previously uncharacterized protein SopA and show that it interacts directly with SgmX, localizes at the leading pole, stimulates polar localization of PilB, and is important for T4aP formation. We corroborate that MglA also recruits FrzS to the leading pole, and FrzS stimulates SgmX recruitment. In addition, FrzS and SgmX separately recruit SopA. Precise quantification of T4aP-formation and T4aP-dependent motility in various mutants supports a model whereby the main pathway for stimulating T4aP formation is the MglA/SgmX pathway. FrzS stimulates this pathway by recruiting SgmX and SopA. SopA stimulates the MglA/SgmX pathway by stimulating the function of SgmX, likely by promoting the SgmX-dependent recruitment of PilB to the T4aP machinery. The architecture of the MglA/SgmX/FrzS/SopA protein interaction network for orchestrating T4aP formation allows for combinatorial regulation of T4aP levels at the leading cell pole resulting in discrete levels of T4aP-dependent motility.
Importance: Type IVa pili (T4aP) are widespread bacterial cell surface structures with important functions in translocation across surfaces, surface adhesion, biofilm formation, and virulence. T4aP-dependent translocation crucially depends on the number of pili. To address how the number of T4aP is regulated, we focused on M. xanthus, which assembles T4aP at the leading cell pole and is a model organism for T4aP biology. Our results support a model whereby the four proteins MglA, SgmX, FrzS, and the newly identified SopA protein establish a highly intricate interaction network for orchestrating T4aP formation at the leading cell pole. This network allows for combinatorial regulation of the number of T4aP resulting in discrete levels of T4aP-dependent motility.
Keywords: FrzS; MglA; Myxococcus xanthus; SgmX; bacterial cell biology; bacterial motility; polarity; type IV pili.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

