Pan-flavivirus analysis reveals sfRNA-independent, 3' UTR-biased siRNA production from an insect-specific flavivirus
- PMID: 39404457
- PMCID: PMC11575252
- DOI: 10.1128/jvi.01215-24
Pan-flavivirus analysis reveals sfRNA-independent, 3' UTR-biased siRNA production from an insect-specific flavivirus
Abstract
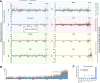
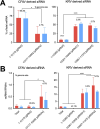
RNA interference (RNAi) plays an essential role in mosquito antiviral immunity, but it is not known whether viral small interfering RNA (siRNA) profiles differ between mosquito-borne and mosquito-specific viruses. A pan-Orthoflavivirus analysis in Aedes albopictus cells revealed that viral siRNAs were evenly distributed across the viral genome of most representatives of the Flavivirus genus. In contrast, siRNA production was biased toward the 3' untranslated region (UTR) of the genomes of classical insect-specific flaviviruses (cISF), which was most pronounced for Kamiti River virus (KRV), a virus with a unique, 1.2 kb long 3' UTR. KRV-derived siRNAs were produced in high quantities and almost exclusively mapped to the 3' UTR. We mapped the 5' end of KRV subgenomic flavivirus RNAs (sfRNAs), products of the 5'-3' exoribonuclease XRN1/Pacman stalling on secondary RNA structures in the 3' UTR of the viral genome. We found that KRV produces high copy numbers of a long, 1,017 nt sfRNA1 and a short, 421 nt sfRNA2, corresponding to two predicted XRN1-resistant elements. Expression of both sfRNA1 and sfRNA2 was reduced in Pacman-deficient Aedes albopictus cells; however, this did not correlate with a shift in viral siRNA profiles. We suggest that cISFs, particularly KRV, developed a unique mechanism to produce high amounts of siRNAs as a decoy for the antiviral RNAi response in an sfRNA-independent manner.IMPORTANCEThe Flavivirus genus contains diverse mosquito viruses ranging from insect-specific viruses circulating exclusively in mosquito populations to mosquito-borne viruses that cause disease in humans and animals. Studying the mechanisms of virus replication and antiviral immunity in mosquitoes is important to understand arbovirus transmission and may inform the development of disease control strategies. In insects, RNA interference (RNAi) provides broad antiviral activity and constitutes a major immune response against viruses. Comparing diverse members of the Flavivirus genus, we found that all flaviviruses are targeted by RNAi. However, the insect-specific Kamiti River virus was unique in that small interfering RNAs are highly skewed toward its uniquely long 3' untranslated region. These results suggest that mosquito-specific viruses have evolved unique mechanisms for genome replication and immune evasion.
Keywords: flavivirus; insect-specific flavivirus; piRNA; siRNA; small RNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Postler TS, Beer M, Blitvich BJ, Bukh J, de Lamballerie X, Drexler JF, Imrie A, Kapoor A, Karganova GG, Lemey P, Lohmann V, Simmonds P, Smith DB, Stapleton JT, Kuhn JH. 2023. Renaming of the genus Flavivirus to Orthoflavivirus and extension of binomial species names within the family Flaviviridae. Arch Virol 168:224. doi: 10.1007/s00705-023-05835-1 - DOI - PubMed
-
- Moureau G, Cook S, Lemey P, Nougairede A, Forrester NL, Khasnatinov M, Charrel RN, Firth AE, Gould EA, de Lamballerie X. 2015. New insights into flavivirus evolution, taxonomy and biogeographic history, extended by analysis of canonical and alternative coding sequences. PLoS One 10:e0117849. doi: 10.1371/journal.pone.0117849 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

