Toxin-Antitoxin Systems Reflect Community Interactions Through Horizontal Gene Transfer
- PMID: 39404847
- PMCID: PMC11523183
- DOI: 10.1093/molbev/msae206
Toxin-Antitoxin Systems Reflect Community Interactions Through Horizontal Gene Transfer
Abstract
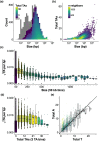
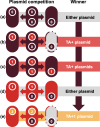
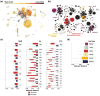
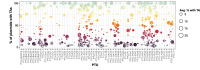
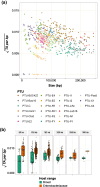
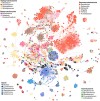
Bacterial evolution through horizontal gene transfer (HGT) reflects their community interactions. In this way, HGT networks do well at mapping community interactions, but offer little toward controlling them-an important step in the translation of synthetic strains into natural contexts. Toxin-antitoxin (TA) systems serve as ubiquitous and diverse agents of selection; however, their utility is limited by their erratic distribution in hosts. Here we examine the heterogeneous distribution of TAs as a consequence of their mobility. By systematically mapping TA systems across a 10,000 plasmid network, we find HGT communities have unique and predictable TA signatures. We propose these TA signatures arise from plasmid competition and have further potential to signal the degree to which plasmids, hosts, and phage interact. To emphasize these relationships, we construct an HGT network based solely on TA similarity, framing specific selection markers in the broader context of bacterial communities. This work both clarifies the evolution of TA systems and unlocks a common framework for manipulating community interactions through TA compatibility.
Keywords: community; horizontal gene transfer; network; plasmid; toxin–antitoxin system.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest The authors declare no competing financial interests.
Figures
References
-
- Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. Proceedings of the international AAAI Conference on Web and Social Media; May 17-20; San Jose, California, USA. p. 3.
MeSH terms
LinkOut - more resources
Full Text Sources

