DeOri 10.0: An Updated Database of Experimentally Identified Eukaryotic Replication Origins
- PMID: 39404857
- PMCID: PMC11652270
- DOI: 10.1093/gpbjnl/qzae076
DeOri 10.0: An Updated Database of Experimentally Identified Eukaryotic Replication Origins
Abstract
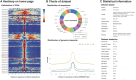
DNA replication is a complex and crucial biological process in eukaryotes. To facilitate the study of eukaryotic replication events, we present a database of eukaryotic DNA replication origins (DeOri), which collects genome-wide data on eukaryotic DNA replication origins currently available. With the rapid development of high-throughput experimental technology in recent years, the number of datasets in the new release of DeOri 10.0 increased from 10 to 151 and the number of sequences increased from 16,145 to 9,742,396. Besides nucleotide sequences and browser extensible data (BED) files, corresponding annotation files, such as coding sequences (CDSs), mRNAs, and other biological elements within replication origins, are also provided. The experimental techniques used for each dataset, as well as related statistical data, are also presented on web page. Differences in experimental methods, cell lines, and sequencing technologies have resulted in distinct replication origins, making it challenging to differentiate between cell-specific and non-specific replication origins. Based on multiple replication origin datasets at the species level, we scored and screened replication origins in Homo sapiens, Gallus gallus, Mus musculus, Drosophila melanogaster, and Caenorhabditis elegans. The screened regions with high scores were considered as species-conservative origins, which are integrated and presented as reference replication origins (rORIs). Additionally, we analyzed the distribution of relevant genomic elements associated with replication origins at the genome level, such as CpG island (CGI), transcription start site (TSS), and G-quadruplex (G4). These analysis results can be browsed and downloaded as needed at http://tubic.tju.edu.cn/deori/.
Keywords: DNA replication; Database; DeOri; Eukaryote; Replication origin.
© The Author(s) 2024. Published by Oxford University Press and Science Press on behalf of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Conflict of interest statement
The authors have declared no competing interests.
Figures


References
-
- Böhly N, Schmidt AK, Zhang X, Slusarenko BO, Hennecke M, Kschischo M, et al. Increased replication origin firing links replication stress to whole chromosomal instability in human cancer. Cell Rep 2022;41:111836. - PubMed
-
- Foss M, McNally FJ, Laurenson P, Rine J.. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 1993;262:1838–44. - PubMed
-
- Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley JF.. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature 1993;366:87–9. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

