Characterisation of the antibody-mediated selective pressure driving intra-host evolution of SARS-CoV-2 in prolonged infection
- PMID: 39405332
- PMCID: PMC11508484
- DOI: 10.1371/journal.ppat.1012624
Characterisation of the antibody-mediated selective pressure driving intra-host evolution of SARS-CoV-2 in prolonged infection
Abstract
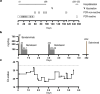
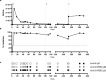
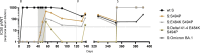
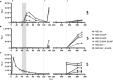
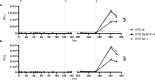
Neutralising antibodies against the SARS-CoV-2 spike (S) protein are major determinants of protective immunity, though insufficient antibody responses may cause the emergence of escape mutants. We studied the humoral immune response causing intra-host evolution in a B-cell depleted, haemato-oncologic patient experiencing clinically severe, prolonged SARS-CoV-2 infection with a virus of lineage B.1.177.81. Following bamlanivimab treatment at an early stage of infection, the patient developed a bamlanivimab-resistant mutation, S:S494P. After five weeks of apparent genetic stability, the emergence of additional substitutions and deletions within the N-terminal domain (NTD) and the receptor binding domain (RBD) of S was observed. Notably, the composition and frequency of escape mutations changed in a short period with an unprecedented dynamic. The triple mutant S:Delta141-4 E484K S494P became dominant until virus elimination. Routine serology revealed no evidence of an antibody response in the patient. A detailed analysis of the variant-specific immune response by pseudotyped virus neutralisation test, surrogate virus neutralisation test, and immunoglobulin-capture enzyme immunoassay showed that the onset of an IgM-dominated antibody response coincided with the appearance of escape mutations. The formation of neutralising antibodies against S:Delta141-4 E484K S494P correlated with virus elimination. One year later, the patient experienced clinically mild re-infection with Omicron BA.1.18, which was treated with sotrovimab and resulted in an increase in Omicron-reactive antibodies. In conclusion, the onset of an IgM-dominated endogenous immune response in an immunocompromised patient coincided with the appearance of additional mutations in the NTD and RBD of S in a bamlanivimab-resistant virus. Although virus elimination was ultimately achieved, this humoral immune response escaped detection by routine diagnosis and created a situation temporarily favouring the rapid emergence of various antibody escape mutants with known epidemiological relevance.
Copyright: © 2024 Schoefbaenker et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

