Generation, expansion, gene delivery, and single-cell profiling in rhesus macaque plasma B cells
- PMID: 39406231
- PMCID: PMC11573788
- DOI: 10.1016/j.crmeth.2024.100878
Generation, expansion, gene delivery, and single-cell profiling in rhesus macaque plasma B cells
Abstract
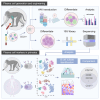
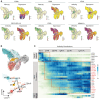
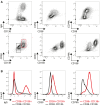
A key step in developing engineered B cells for therapeutic purposes is evaluation in immunocompetent, large-animal models. Therefore, we developed methods to purify, expand, and differentiate non-human primate (NHP; rhesus macaque) B cells. After 7 days in culture, B cells expanded 10-fold, differentiated into a plasma cell phenotype (CD38, CD138), and secreted immunoglobulin G. Using single-cell sequencing and flow cytometry, we verified the presence of plasma cell genes in differentiated NHP B cells and unearthed less-recognized markers, such as CD59 and CD79A. In contrast with human cells, we found that the immune checkpoint molecule CD274 (PD-L1) and major histocompatibility complex (MHC) class I molecules were upregulated in NHP plasma cells in the transcriptional data. Lastly, we established the conditions for efficient transduction of NHP B cells with adeno-associated virus (AAV) vectors, achieving a delivery rate of approximately 60%. We envision that this work will accelerate proof-of-concept studies using engineered B cells in NHPs.
Keywords: AAV; CD59; CD79A; CP: Biotechnology; CP: Immunology; MHC class I; NHP; PD-L1; cell therapy; differentiation; expansion; plasma cell; primate.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.G.J. and D.J.R. hold equity in and serve on the scientific advisory board of Be Biopharma, Inc. S.K. is currently an employee at Astellas Pharma, Y.H. is currently an employee of Sonoma Biotherapeutics, and C.L. is currently an employee of Sartorius AG.
Figures





References
-
- Vamva E., Ozog S., Leaman D.P., Yu-Hong Cheng R., Irons N.J., Ott A., Stoffers C., Khan I., Goebrecht G.K.E., Gardner M.R., et al. A lentiviral vector B cell gene therapy platform for the delivery of the anti-HIV-1 eCD4-Ig-knob-in-hole-reversed immunoadhesin. Mol. Ther. Methods Clin. Dev. 2023;28:366–384. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

