Complex structural variation is prevalent and highly pathogenic in pediatric solid tumors
- PMID: 39406233
- PMCID: PMC11605687
- DOI: 10.1016/j.xgen.2024.100675
Complex structural variation is prevalent and highly pathogenic in pediatric solid tumors
Abstract
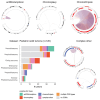
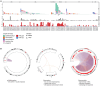
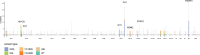
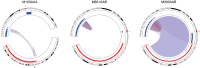
In pediatric cancer, structural variants (SVs) and copy-number alterations contribute to cancer initiation as well as progression, thereby aiding diagnosis and treatment stratification. Although suggested to be of importance, the prevalence and biological relevance of complex genomic rearrangements (CGRs) across pediatric solid tumors is largely unexplored. In a cohort of 120 primary tumors, we systematically characterized patterns of extrachromosomal DNA, chromoplexy, and chromothripsis across five pediatric solid cancer types. CGRs were identified in 56 tumors (47%), and in 42 of these tumors, CGRs affect cancer driver genes or result in unfavorable chromosomal alterations. This demonstrates that CGRs are prevalent and pathogenic in pediatric solid tumors and suggests that selection likely contributes to the structural variation landscape. Moreover, carrying CGRs is associated with more adverse clinical events. Our study highlights the potential for CGRs to be incorporated in risk stratification or exploited for targeted treatments.
Keywords: CGRs; WGS; chromoplexy; chromothripsis; complex genomic rearrangements; complex structural variation; ecDNA; extrachromosomal DNA; pediatric solid tumors; whole-genome sequencing.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

