Genome-wide association studies in a diverse strawberry collection unveil loci controlling agronomic and fruit quality traits
- PMID: 39406253
- PMCID: PMC11628880
- DOI: 10.1002/tpg2.20509
Genome-wide association studies in a diverse strawberry collection unveil loci controlling agronomic and fruit quality traits
Abstract
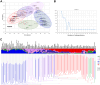
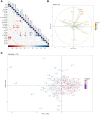
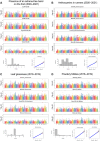
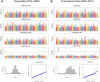
Strawberries (Fragaria sp.) are cherished for their organoleptic properties and nutritional value. However, breeding new cultivars involves the simultaneous selection of many agronomic and fruit quality traits, including fruit firmness and extended postharvest life. The strawberry germplasm collection here studied exhibited extensive phenotypic variation in 26 agronomic and fruit quality traits across three consecutive seasons. Phenotypic correlations and principal component analysis revealed relationships among traits and accessions, emphasizing the impact of plant breeding on fruit weight and firmness to the detriment of sugar or vitamin C content. Genetic diversity analysis on 124 accessions using 44,408 markers denoted a population structure divided into six subpopulations still retaining considerable diversity. Genome-wide association studies for the 26 traits unveiled 121 significant marker-trait associations distributed across 95 quantitative trait loci (QTLs). Multiple associations were detected for fruit firmness, a key breeding target, including a prominent locus on chromosome 6A. The candidate gene FaPG1, controlling fruit softening and postharvest shelf life, was identified within this QTL region. Differential expression of FaPG1 confirmed its role as the primary contributor to natural variation in fruit firmness. A kompetitive allele-specific PCR assay based on the single nucleotide polymorphism (SNP) AX-184242253, associated with the 6A QTL, predicts a substantial increase in fruit firmness, validating its utility for marker-assisted selection. In essence, this comprehensive study provides insights into the phenotypic and genetic landscape of the strawberry collection and lays a robust foundation for propelling the development of superior strawberry cultivars through precision breeding.
© 2024 The Author(s). The Plant Genome published by Wiley Periodicals LLC on behalf of Crop Science Society of America.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Antanaviciute, L. , Vickerstaff, R. , Gomez‐Cortecero, A. , Marina‐Montes, C. , Llorente‐Garcia, A. , Battey, N. H. , & Harrison, R. J. (2017). Correlation analysis and QTL mapping of fruit quality and plant architecture traits in cultivated strawberry (Fragaria × ananassa). Acta Horticulturae, 1172, 307–316. 10.17660/ActaHortic.2017.1172.58 - DOI
-
- Atwell, S. , Huang, Y. S. , Vilhjálmsson, B. J. , Willems, G. , Horton, M. , Li, Y. , Meng, D. , Platt, A. , Tarone, A. M. , Hu, T. T. , Jiang, R. , Muliyati, N. W. , Zhang, X. , Amer, M. A. , Baxter, I. , Brachi, B. , Chory, J. , Dean, C. , Debieu, M. , … Nordborg, M. (2010). Genome‐wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature, 465, 627–631. 10.1038/nature08800 - DOI - PMC - PubMed
-
- Barbey, C. R. , Hogshead, M. H. , Harrison, B. , Schwartz, A. E. , Verma, S. , Oh, Y. , Lee, S. , Folta, K. M. , & Whitaker, V. M. (2021). Genetic analysis of methyl anthranilate, mesifurane, linalool, and other flavor compounds in cultivated strawberry (Fragaria × ananassa) . Frontiers in Plant Science, 12, Article 615749. 10.3389/fpls.2021.615749 - DOI - PMC - PubMed
-
- Bassil, N. V. , Davis, T. M. , Zhang, H. , Ficklin, S. , Mittmann, M. , Webster, T. , Mahoney, L. , Wood, D. , Alperin, E. S. , Rosyara, U. R. , Koehorst‐vanc Putten, H. , Monfort, A. , Sargent, D. J. , Amaya, I. , Denoyes, B. , Bianco, L. , van Dijk, T. , Pirani, A. , Iezzoni, A. , … van de Weg, E. (2015). Development and preliminary evaluation of a 90 K Axiom® SNP array for the allo‐octoploid cultivated strawberry Fragaria × ananassa . BMC Genomics, 16(155), Article 155. 10.1186/s12864-015-1310-1 - DOI - PMC - PubMed
MeSH terms
Grants and funding
- P18-RT-4856/Consejería de Economía, Innovación, Ciencia y Empleo, Junta de Andalucía
- BreedingValue 101000747/Horizon 2020 Framework Programme
- PID2022-138290OR-I00/AEI/10.13039/501100011033/Ministerio de Ciencia e Innovación y Agencia Estatal de Investigación
- PID2019-111496RR-I00/AEI/10.13039/501100011033/FEDER, UE Ministerio de Ciencia e Innovación y Agencia Estatal de Investigación
LinkOut - more resources
Full Text Sources

