Genetic and evolutionary divergence of harbour seals (Phoca vitulina) in Iliamna Lake, Alaska
- PMID: 39406337
- PMCID: PMC11523099
- DOI: 10.1098/rsbl.2024.0166
Genetic and evolutionary divergence of harbour seals (Phoca vitulina) in Iliamna Lake, Alaska
Abstract
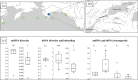
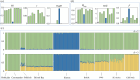
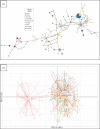
Freshwater populations of typically marine species present unique opportunities to investigate biodiversity, evolutionary divergence, and the adaptive potential and niche width of species. A few pinniped species have populations that reside solely in freshwater. The harbour seals inhabiting Iliamna Lake, Alaska constitute one such population. Their remoteness, however, has long hindered scientific inquiry. We used DNA from seal scat and tissue samples provided by Indigenous hunters to screen for mitochondrial DNA and microsatellite variation within Iliamna Lake and eight regions across the Pacific Ocean. The Iliamna seals (i) were substantially and significantly discrete from all other populations ( [Formula: see text]F st-mtDNA = 0.544, [Formula: see text]Φ st - mtDNA = 0.541, [Formula: see text]F st-microsatellites = 0.308), (ii) formed a discrete genetic cluster separate from all marine populations (modal ∆k = 2, PC1 = 14.8%), had (iii) less genetic diversity (Hd, π, H exp), and (iv) higher inbreeding (F) than marine populations. These findings are both striking and unexpected revealing that Iliamna seals have likely been on a separate evolutionary trajectory for some time and may represent a unique evolutionary legacy for the species. Attention must now be given to the selective processes driving evolutionary divergence from harbour seals in marine habitats and to ensuring the future of the Iliamna seal.
Keywords: Iliamna Lake; Phoca vitulina; divergences; evolution; genetics; seals.
Conflict of interest statement
We declare we have no competing interests.
Figures
References
-
- Burns J, et al. . 2016. Integrating local traditional knowledge and subsistence use patterns with aerial surveys to improve scientific and local understanding of the iliamna lake seals. Technical paper no. 416. Juneau, AK: Alaska Department of Fish and Game, Division of Subsistence.
-
- Burns J, Withrow D, Van Lanen JM. 2018. Freshwater seals of Iliamna Lake. In Bristol Bay Alaska: natural resources of the aquatic and terrestrial ecosystems (ed. Woody CA), pp. 493–508. Plantation, FL: J. Ross Publishing.
-
- Würsig B, Thewissen JGM, Kovacs KM. 2018. Encyclopedia of marine mammals. San Diego, CA: Academic Press.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
