Targeting IRE1α reprograms the tumor microenvironment and enhances anti-tumor immunity in prostate cancer
- PMID: 39406723
- PMCID: PMC11480464
- DOI: 10.1038/s41467-024-53039-1
Targeting IRE1α reprograms the tumor microenvironment and enhances anti-tumor immunity in prostate cancer
Abstract
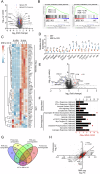
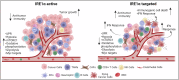
Unfolded protein response (UPR) is a central stress response pathway that is hijacked by tumor cells for their survival. Here, we find that IRE1α signaling, one of the canonical UPR arms, is increased in prostate cancer (PCa) patient tumors. Genetic or small molecule inhibition of IRE1α in syngeneic mouse PCa models and an orthotopic model decreases tumor growth. IRE1α ablation in cancer cells potentiates interferon responses and activates immune system related pathways in the tumor microenvironment (TME). Single-cell RNA-sequencing analysis reveals that targeting IRE1α in cancer cells reduces tumor-associated macrophage abundance. Consistently, the small molecule IRE1α inhibitor MKC8866, currently in clinical trials, reprograms the TME and enhances anti-PD-1 therapy. Our findings show that IRE1α signaling not only promotes cancer cell growth and survival but also interferes with anti-tumor immunity in the TME. Thus, targeting IRE1α can be a promising approach for improving anti-PD-1 immunotherapy in PCa.
© 2024. The Author(s).
Conflict of interest statement
JBP is employee and shareholder of Fosun Orinove. The remaining authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- 303353/Norges Forskningsråd (Research Council of Norway)
- 313932/Norges Forskningsråd (Research Council of Norway)
- 247110/Kreftforeningen (Norwegian Cancer Society)
- 214871/Kreftforeningen (Norwegian Cancer Society)
- 102583101/Ministry of Health and Care Services | Helse Sør-Øst RHF (Southern and Eastern Norway Regional Health Authority)
- 10000/Anders Jahres Fond til Vitenskapens Fremme (Anders Jahre's Foundation for the Promotion of Science)
- 1109-UBC GR026025/Terry Fox Foundation
- 1109-UBC GR026025/Terry Fox Foundation
- 311-A18039/Kræftens Bekæmpelse (Danish Cancer Society)
- 187615/University of Oslo | Livsvitenskap, Universitetet i Oslo (UiO:Life Science)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

