A genome-first study of sex chromosome aneuploidies provides evidence of Y chromosome dosage effects on autism risk
- PMID: 39406744
- PMCID: PMC11480344
- DOI: 10.1038/s41467-024-53211-7
A genome-first study of sex chromosome aneuploidies provides evidence of Y chromosome dosage effects on autism risk
Abstract
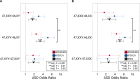
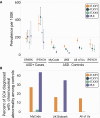
A female protective effect has long been postulated as the primary explanation for the four-fold increase of autism spectrum disorder (ASD) diagnoses in males versus females. However, genetic and epidemiological investigations of this hypothesis have so far failed to explain the large difference in ASD prevalence between the sexes. To address this knowledge gap, we examined sex chromosome aneuploidy in a large ASD case-control cohort to evaluate the relationship between X and Y chromosome dosage and ASD risk. From these data, we modeled three relationships between sex chromosome dosage and ASD risk: the extra Y effect, the extra X effect, and sex chromosome haploinsufficiency. We found that the extra Y effect increased ASD risk significantly more than the extra X effect. Among females, we observed a large association between 45, X and ASD, confirming sex chromosome haploinsufficiency as a strong ASD risk factor. These results provide a framework for understanding the relationship between X and Y chromosome dosage on ASD, which may inform future research investigating genomic contributors to the observed sex difference.
© 2024. The Author(s).
Conflict of interest statement
D.H. Ledbetter reported being a scientific consultant to Nest Genomics, MyOme, Singular Genomics, and X-Therma. No other disclosures were reported.
Figures



References
-
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition. (American Psychiatric Publishing, Arlington, VA, 2013).
-
- Falconer, D. S. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann. Hum. Genet.29, 51–76 (1965). - DOI
Publication types
MeSH terms
Grants and funding
- R01 MH074090/MH/NIMH NIH HHS/United States
- OT2 OD026556/OD/NIH HHS/United States
- U2C OD023196/OD/NIH HHS/United States
- OT2 OD025315/OD/NIH HHS/United States
- OT2 OD026551/OD/NIH HHS/United States
- OT2 OD026552/OD/NIH HHS/United States
- OT2 OD026549/OD/NIH HHS/United States
- OT2 OD025337/OD/NIH HHS/United States
- OT2 OD025277/OD/NIH HHS/United States
- OT2 OD026555/OD/NIH HHS/United States
- OT2 OD026550/OD/NIH HHS/United States
- OT2 OD026553/OD/NIH HHS/United States
- OT2 OD023205/OD/NIH HHS/United States
- OT2 OD025276/OD/NIH HHS/United States
- OT2 OD026557/OD/NIH HHS/United States
- OT2 OD026554/OD/NIH HHS/United States
- U24 OD023163/OD/NIH HHS/United States
- OT2 OD023206/OD/NIH HHS/United States
- U24 OD023176/OD/NIH HHS/United States
- OT2 OD026548/OD/NIH HHS/United States
- U24 OD023121/OD/NIH HHS/United States
- U01 MH119705/MH/NIMH NIH HHS/United States
- AOD16027001/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

