Exome variant prioritization in a large cohort of hearing-impaired individuals indicates IKZF2 to be associated with non-syndromic hearing loss and guides future research of unsolved cases
- PMID: 39406892
- PMCID: PMC11522133
- DOI: 10.1007/s00439-024-02706-w
Exome variant prioritization in a large cohort of hearing-impaired individuals indicates IKZF2 to be associated with non-syndromic hearing loss and guides future research of unsolved cases
Abstract
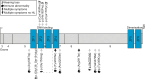
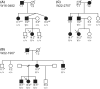
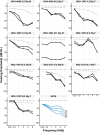
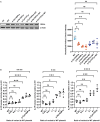
Although more than 140 genes have been associated with non-syndromic hereditary hearing loss (HL), at least half of the cases remain unexplained in medical genetic testing. One reason is that pathogenic variants are located in 'novel' deafness genes. A variant prioritization approach was used to identify novel (candidate) genes for HL. Exome-wide sequencing data were assessed for subjects with presumed hereditary HL that remained unexplained in medical genetic testing by gene-panel analysis. Cases in group AD had presumed autosomal dominantly inherited HL (n = 124), and in group AR, presumed autosomal recessive HL (n = 337). Variants in known and candidate deafness genes were prioritized based on allele frequencies and predicted effects. Selected variants were tested for their co-segregation with HL. Two cases were solved by variants in recently identified deafness genes (ABHD12, TRRAP). Variant prioritization also revealed potentially causative variants in candidate genes associated with recessive and X-linked HL. Importantly, missense variants in IKZF2 were found to co-segregate with dominantly inherited non-syndromic HL in three families. These variants specifically affected Zn2+-coordinating cysteine or histidine residues of the zinc finger motifs 2 and 3 of the encoded protein Helios. This finding indicates a complex genotype-phenotype correlation for IKZF2 defects, as this gene was previously associated with non-syndromic dysfunction of the immune system and ICHAD syndrome, including HL. The designed strategy for variant prioritization revealed that IKZF2 variants can underlie non-syndromic HL. The large number of candidate genes for HL and variants therein stress the importance of inclusion of family members for variant prioritization.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures


References
-
- Baldarelli RM, Smith CM, Finger JH, Hayamizu TF, McCright IJ, Xu J, Shaw DR, Beal JS, Blodgett O, Campbell J, Corbani LE, Frost PJ, Giannatto SC, Miers DB, Kadin JA, Richardson JE, Ringwald M (2021) The mouse Gene Expression Database (GXD): 2021 update. Nucleic Acids Res 49(D1):D924-d931. 10.1093/nar/gkaa914 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

