Transcriptome and acetylome profiling identify crucial steps of neuronal differentiation in Rubinstein-Taybi syndrome
- PMID: 39407026
- PMCID: PMC11480426
- DOI: 10.1038/s42003-024-06939-3
Transcriptome and acetylome profiling identify crucial steps of neuronal differentiation in Rubinstein-Taybi syndrome
Abstract
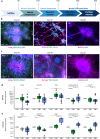
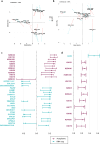
Rubinstein-Taybi syndrome (RTS) is a rare and severe genetic developmental disorder characterized by multiple congenital anomalies and intellectual disability. CREBBP and EP300, the two genes known to cause RTS encode transcriptional coactivators with a catalytic lysine acetyltransferase (KAT) activity. Loss of CBP or p300 function results in a deficit in protein acetylation, in particular at histones. In RTS, nothing is known on the consequences of the loss of histone acetylation on the transcriptomic profiles during neuronal differentiation. To address this question, we differentiated induced pluripotent stem cells from RTS patients carrying a recurrent CREBBP mutation that inactivates the KAT domain into cortical and pyramidal neurons. By comparing their acetylome and their transcriptome at different neuronal differentiation time points, we identified 25 specific acetylated histone residues altered in RTS. We also identified the transition between neural progenitors and immature neurons as a critical step of the differentiation process, with a delayed neuronal maturation in RTS. Overall, this study opens new perspectives in the definition of epigenetic biomarkers for RTS, whose methodology could be extended to other chromatinopathies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

