Identification of sex-specific markers using genome re-sequencing in the blunt snout bream (Megalobrama amblycephala)
- PMID: 39407110
- PMCID: PMC11481317
- DOI: 10.1186/s12864-024-10884-0
Identification of sex-specific markers using genome re-sequencing in the blunt snout bream (Megalobrama amblycephala)
Abstract
Background: The blunt snout bream (Megalobrama amblycephala) is an important economic freshwater fish in China with tender flesh and high nutritional value. With the cultivation of superior new varieties and the expansion of breeding scale, it becomes imperative to employ sex-control technology to cultivate monosexual populations of M. amblycephala, thereby preventing the deterioration of desirable traits. The development of specific markers capable of accurately identifying the sex of M. amblycephala would facilitate the determination of the genetic sex of the breeding population before gonad maturation, thereby expediting the processes of sex-controlled breeding of M. amblycephala.
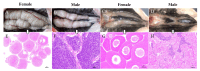
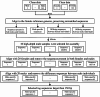
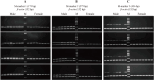
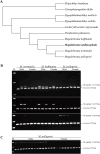
Results: A whole-genome re-sequencing was performed for 116 females and 141 males M. amblycephala collected from nine populations. Seven candidate male-specific sequences were identified through comparative analysis of male and female genomes, which were further compared with the sequencing data of 257 individuals, and finally three male-specific sequences were generated. These three sequences were further validated by PCR amplification in 32 males and 32 females to confirm their potential as male-specific molecular markers for M. amblycephala. One of these markers showed potential applicability in M. pellegrini as well, enabling males to be identified using this specific molecular marker.
Conclusions: The study provides a high-efficiency and cost-effective approach for the genetic sex identification in two species of Megalobrama. The developed markers in this study have great potential in facilitating sex-controlled breeding of M. amblycephala and M. pellegrini, while also contributing valuable insights into the underlying mechanisms of fish sex determination.
Keywords: Megalobrama amblycephala; Genetic sex identification; Molecular marker; Whole-genome re-sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Transcriptome analysis and SSR/SNP markers information of the blunt snout bream (Megalobrama amblycephala).PLoS One. 2012;7(8):e42637. doi: 10.1371/journal.pone.0042637. Epub 2012 Aug 6. PLoS One. 2012. PMID: 22880060 Free PMC article.
-
An AFLP-based approach for the identification of sex-linked markers in blunt snout bream, Megalobrama amblycephala (Cyprinidae).Genet Mol Res. 2012 Apr 19;11(2):1027-31. doi: 10.4238/2012.April.19.7. Genet Mol Res. 2012. PMID: 22576928
-
Genetic diversity of and differentiation among five populations of blunt snout bream (Megalobrama amblycephala) revealed by SRAP markers: implications for conservation and management.PLoS One. 2014 Sep 29;9(9):e108967. doi: 10.1371/journal.pone.0108967. eCollection 2014. PLoS One. 2014. PMID: 25265288 Free PMC article.
-
Identification and characterization of microRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by Solexa sequencing.BMC Genomics. 2013 Nov 5;14:754. doi: 10.1186/1471-2164-14-754. BMC Genomics. 2013. PMID: 24188211 Free PMC article.
-
Is the Nutritional Value of Fish Fillet Related to Fish Maturation or Fish Age? Integrated Analysis of Transcriptomics and Metabolomics in Blunt Snout Bream (Megalobrama amblycephala).Cell Physiol Biochem. 2018;49(1):17-39. doi: 10.1159/000492837. Epub 2018 Aug 22. Cell Physiol Biochem. 2018. PMID: 30134220
References
-
- Miya M, Gotoh RO, Sado T. MiFish metabarcoding: a high-throughput approach for simultaneous detection of multiple fish species from environmental DNA and other samples. Fisheries Sci. 2020;86:939–70.
-
- Chen J, Hu W, Zhu ZY. Progress in studies of fish reproductive development regulation. Chin Sci Bull. 2013;58:7–16.
-
- Li XY, Gui JF. Diverse and variable sex determination mechanisms in vertebrates. Sci China Life Sci. 2018;61:1503–14. - PubMed
-
- Kobayashi Y, Nagahama Y, Nakamura M. Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev. 2013;7:115–25. - PubMed
-
- Mei J, Gui JF. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci China Life Sci. 2015;58:124–36. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

