Differential methylation of circulating free DNA assessed through cfMeDiP as a new tool for breast cancer diagnosis and detection of BRCA1/2 mutation
- PMID: 39407254
- PMCID: PMC11476115
- DOI: 10.1186/s12967-024-05734-2
Differential methylation of circulating free DNA assessed through cfMeDiP as a new tool for breast cancer diagnosis and detection of BRCA1/2 mutation
Abstract
Background: Recent studies have highlighted the importance of the cell-free DNA (cfDNA) methylation profile in detecting breast cancer (BC) and its different subtypes. We investigated whether plasma cfDNA methylation, using cell-free Methylated DNA Immunoprecipitation and High-Throughput Sequencing (cfMeDIP-seq), may be informative in characterizing breast cancer in patients with BRCA1/2 germline mutations for early cancer detection and response to therapy.
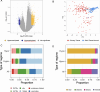
Methods: We enrolled 23 BC patients with germline mutation of BRCA1 and BRCA2 genes, 19 healthy controls without BRCA1/2 mutation, and two healthy individuals who carried BRCA1/2 mutations. Blood samples were collected for all study subjects at the diagnosis, and plasma was isolated by centrifugation. Cell-free DNA was extracted from 1 mL of plasma, and cfMeDIP-seq was performed for each sample. Shallow whole genome sequencing was performed on the immuno-precipitated samples. Then, the differentially methylated 300-bp regions (DMRs) between 25 BRCA germline mutation carriers and 19 non-carriers were identified. DMRs were compared with tumor-specific regions from public datasets to perform an unbiased analysis. Finally, two statistical classifiers were trained based on the GLMnet and random forest model to evaluate if the identified DMRs could discriminate BRCA-positive from healthy samples.
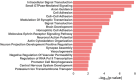
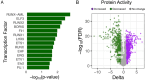
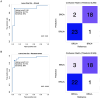
Results: We identified 7,095 hypermethylated and 212 hypomethylated regions in 25 BRCA germline mutation carriers compared to 19 controls. These regions discriminate tumors from healthy samples with high accuracy and sensitivity. We show that the circulating tumor DNA of BRCA1/2 mutant breast cancers is characterized by the hypomethylation of genes involved in DNA repair and cell cycle. We uncovered the TFs associated with these DRMs and identified that proteins of the Erythroblast Transformation Specific (ETS) family are particularly active in the hypermethylated regions. Finally, we assessed that these regions could discriminate between BRCA positives from healthy samples with an AUC of 0.95, a sensitivity of 88%, and a specificity of 94.74%.
Conclusions: Our study emphasizes the importance of tumor cell-derived DNA methylation in BC, reporting a different methylation profile between patients carrying mutations in BRCA1, BRCA2, and wild-type controls. Our minimally invasive approach could allow early cancer diagnosis, assessment of minimal residual disease, and monitoring of response to therapy.
Keywords: BRCA1; BRCA2; Breast cancer; Cell-free DNA; DMRs; cfMeDIP-seq.
© 2024. The Author(s).
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures
References
-
- Prat A, Pineda E, Adamo B, Galván P, Fernández A, Gaba L, et al. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl 2):S26–35. - PubMed
-
- Wan A, Zhang G, Ma D, Zhang Y, Qi X. An overview of the research progress of BRCA gene mutations in breast cancer. Biochim Biophys Acta Rev Cancer. 2023;1878(4):188907. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

