Characterization of Circulating Protein Profiles in Individuals with Prader-Willi Syndrome and Individuals with Non-Syndromic Obesity
- PMID: 39407757
- PMCID: PMC11476631
- DOI: 10.3390/jcm13195697
Characterization of Circulating Protein Profiles in Individuals with Prader-Willi Syndrome and Individuals with Non-Syndromic Obesity
Abstract
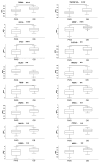
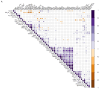
Background: Prader-Willi syndrome (PWS) is a rare genetic disorder characterized by distinctive physical, cognitive, and behavioral manifestations, coupled with profound alterations in appetite regulation, leading to severe obesity and metabolic dysregulation. These clinical features arise from disruptions in neurodevelopment and neuroendocrine regulation, yet the molecular intricacies of PWS remain incompletely understood. Methods: This study aimed to comprehensively profile circulating neuromodulatory factors in the serum of 53 subjects with PWS and 34 patients with non-syndromic obesity, utilizing a proximity extension assay with the Olink Target 96 neuro-exploratory and neurology panels. The ANOVA p-values were adjusted for multiple testing using the Benjamani-Hochberg method. Protein-protein interaction networks were generated in STRING V.12. Corrplots were calculated with R4.2.2 by using the Hmisc, Performance Analytics, and Corrplot packages Results: Our investigation explored the potential genetic underpinnings of the circulating protein signature observed in PWS, revealing intricate connections between genes in the PWS critical region and the identified circulating proteins associated with impaired oxytocin, NAD metabolism, and sex-related neuromuscular impairment involving, CD38, KYNU, NPM1, NMNAT1, WFIKKN1, and GDF-8/MSTN. The downregulation of CD38 in individuals with PWS (p < 0.01) indicates dysregulation of oxytocin release, implicating pathways associated with NAD metabolism in which KYNU and NMNAT1 are involved and significantly downregulated in PWS (p < 0.01 and p < 0.05, respectively). Sex-related differences in the circulatory levels of WFIKKN1 and GDF-8/MSTN (p < 0.05) were also observed. Conclusions: This study highlights potential circulating protein biomarkers associated with impaired oxytocin, NAD metabolism, and sex-related neuromuscular impairment in PWS individuals with potential clinical implications.
Keywords: Prader–Willi syndrome; circulating biomarkers; neuromodulatory factors; non-syndromic obesity; proteome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Butler M.G., Hartin S.N., Hossain W.A., Manzardo A.M., Kimonis V., Dykens E., Gold J.A., Kim S.-J., Weisensel N., Tamura R., et al. Molecular Genetic Classification in Prader-Willi Syndrome: A Multisite Cohort Study. J. Med. Genet. 2019;56:149–153. doi: 10.1136/jmedgenet-2018-105301. - DOI - PMC - PubMed
-
- Pacoricona Alfaro D.L., Lemoine P., Ehlinger V., Molinas C., Diene G., Valette M., Pinto G., Coupaye M., Poitou-Bernert C., Thuilleaux D., et al. Causes of Death in Prader-Willi Syndrome: Lessons from 11 Years’ Experience of a National Reference Center. Orphanet J. Rare Dis. 2019;14:238. doi: 10.1186/s13023-019-1214-2. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

