Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage
- PMID: 39408633
- PMCID: PMC11477204
- DOI: 10.3390/ijms251910304
Predicting Outcomes of Preterm Neonates Post Intraventricular Hemorrhage
Abstract
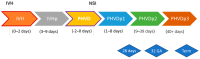
Intraventricular hemorrhage (IVH) in preterm neonates presents a high risk for developing posthemorrhagic ventricular dilatation (PHVD), a severe complication that can impact survival and long-term outcomes. Early detection of PHVD before clinical onset is crucial for optimizing therapeutic interventions and providing accurate parental counseling. This study explores the potential of explainable machine learning models based on targeted liquid biopsy proteomics data to predict outcomes in preterm neonates with IVH. In recent years, research has focused on leveraging advanced proteomic technologies and machine learning to improve prediction of neonatal complications, particularly in relation to neurological outcomes. Machine learning (ML) approaches, combined with proteomics, offer a powerful tool to identify biomarkers and predict patient-specific risks. However, challenges remain in integrating large-scale, multiomic datasets and translating these findings into actionable clinical tools. Identifying reliable, disease-specific biomarkers and developing explainable ML models that clinicians can trust and understand are key barriers to widespread clinical adoption. In this prospective longitudinal cohort study, we analyzed 1109 liquid biopsy samples from 99 preterm neonates with IVH, collected at up to six timepoints over 13 years. Various explainable ML techniques-including statistical, regularization, deep learning, decision trees, and Bayesian methods-were employed to predict PHVD development and survival and to discover disease-specific protein biomarkers. Targeted proteomic analyses were conducted using serum and urine samples through a proximity extension assay capable of detecting low-concentration proteins in complex biofluids. The study identified 41 significant independent protein markers in the 1600 calculated ML models that surpassed our rigorous threshold (AUC-ROC of ≥0.7, sensitivity ≥ 0.6, and selectivity ≥ 0.6), alongside gestational age at birth, as predictive of PHVD development and survival. Both known biomarkers, such as neurofilament light chain (NEFL), and novel biomarkers were revealed. These findings underscore the potential of targeted proteomics combined with ML to enhance clinical decision-making and parental counseling, though further validation is required before clinical implementation.
Keywords: biomarker; intensive care; intraventricular hemorrhage; machine learning; neonate; posthemorrhagic hydrocephalus; prediction; prematurity; proteomics; survival.
Conflict of interest statement
Authors Gabriel A. Vignolle, Priska Bauerstätter, Silvia Schönthaler and Christa Nöhammer were employed by the company AIT Austrian Institute of Technology GmbHThe. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Leijser L.M., Miller S.P., van Wezel-Meijler G., Brouwer A.J., Traubici J., van Haastert I.C., Whyte H.E., Groenendaal F., Kulkarni A.V., Han K.S., et al. Posthemorrhagic ventricular dilatation in preterm infants: When best to intervene? Neurology. 2018;90:e698–e706. doi: 10.1212/WNL.0000000000004984. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

