Profiling Reduced Expression of Contractile and Mitochondrial mRNAs in the Human Sinoatrial Node vs. Right Atrium and Predicting Their Suppressed Expression by Transcription Factors and/or microRNAs
- PMID: 39408732
- PMCID: PMC11477614
- DOI: 10.3390/ijms251910402
Profiling Reduced Expression of Contractile and Mitochondrial mRNAs in the Human Sinoatrial Node vs. Right Atrium and Predicting Their Suppressed Expression by Transcription Factors and/or microRNAs
Abstract
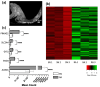
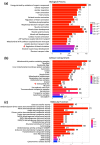
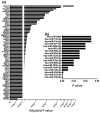
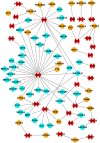
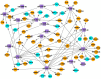
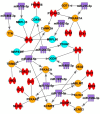
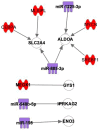
(1) Background: The sinus node (SN) is the main pacemaker of the heart. It is characterized by pacemaker cells that lack mitochondria and contractile elements. We investigated the possibility that transcription factors (TFs) and microRNAs (miRs) present in the SN can regulate gene expression that affects SN morphology and function. (2) Methods: From human next-generation sequencing data, a list of mRNAs that are expressed at lower levels in the SN compared with the right atrium (RA) was compiled. The mRNAs were then classified into contractile, mitochondrial or glycogen mRNAs using bioinformatic software, RStudio and Ingenuity Pathway Analysis. The mRNAs were combined with TFs and miRs to predict their interactions. (3) Results: From a compilation of the 1357 mRNAs, 280 contractile mRNAs and 198 mitochondrial mRNAs were identified to be expressed at lower levels in the SN compared with RA. TFs and miRs were shown to interact with contractile and mitochondrial function-related mRNAs. (4) Conclusions: In human SN, TFs (MYCN, SOX2, NUPR1 and PRDM16) mainly regulate mitochondrial mRNAs (COX5A, SLC25A11 and NDUFA8), while miRs (miR-153-3p, miR-654-5p, miR-10a-5p and miR-215-5p) mainly regulate contractile mRNAs (RYR2, CAMK2A and PRKAR1A). TF and miR-mRNA interactions provide a further understanding of the complex molecular makeup of the SN and potential therapeutic targets for cardiovascular treatments.
Keywords: Ingenuity Pathway Analysis; contractile function; glycogen metabolism; human right atrium; human sinus node/sinoatrial node; miRNA; mitochondrial function; transcription factor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Stephenson R.S., Atkinson A., Kottas P., Perde F., Jafarzadeh F., Bateman M., Iaizzo P.A., Zhao J., Zhang H., Anderson R.H., et al. High resolution 3-Dimensional imaging of the human cardiac conduction system from microanatomy to mathematical modeling. Sci. Rep. 2017;7:7188. doi: 10.1038/s41598-017-07694-8. - DOI - PMC - PubMed
-
- Dobrzynski H., Anderson R.H., Atkinson A., Borbas Z., D’Souza A., Fraser J.F., Inada S., Logantha S.J., Monfredi O., Morris G.M., et al. Structure, function and clinical relevance of the cardiac conduction system, including the atrioventricular ring and outflow tract tissues. Pharmacol. Ther. 2013;139:260–288. doi: 10.1016/j.pharmthera.2013.04.010. - DOI - PubMed
-
- Chandler N.J., Greener I.D., Tellez J.O., Inada S., Musa H., Molenaar P., Difrancesco D., Baruscotti M., Longhi R., Anderson R.H., et al. Molecular architecture of the human sinus node: Insights into the function of the cardiac pacemaker. Circulation. 2009;119:1562–1575. doi: 10.1161/CIRCULATIONAHA.108.804369. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

