Characterization of Endothelial Cell Subclusters in Localized Scleroderma Skin with Single-Cell RNA Sequencing Identifies NOTCH Signaling Pathway
- PMID: 39408800
- PMCID: PMC11477421
- DOI: 10.3390/ijms251910473
Characterization of Endothelial Cell Subclusters in Localized Scleroderma Skin with Single-Cell RNA Sequencing Identifies NOTCH Signaling Pathway
Abstract
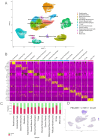
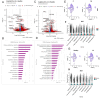
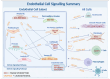
Localized scleroderma (LS) is an autoimmune disease characterized by inflammation and fibrosis, leading to severe cutaneous manifestations such as skin hardening, tightness, discoloration, and other textural changes that may result in disability. While LS shares similar histopathologic features and immune-fibroblast interactions with systemic sclerosis (SSc), its molecular mechanisms remain understudied. Endothelial cells (EC) are known to play a crucial role in SSc but have not been investigated in LS. Single-cell RNA sequencing (scRNA-seq) now allows for detailed examination of this cell type in the primary organ of interest for scleroderma, the skin. In this study, we analyzed skin-isolated cells from 27 LS patients (pediatric and adult) and 17 healthy controls using scRNA-seq. Given the known role of EC damage as an initial event in SSc and the histologic and clinical skin similarities to LS, we focused primarily on endothelial cells. Our analysis identified eight endothelial subclusters within the dataset, encompassing both disease and healthy samples. Interaction analysis revealed that signaling from diseased endothelial cells was predicted to promote fibrosis through SELE interaction with FGFBP1 and other target genes. We also observed high levels of JAG in arterial endothelial cells and NOTCH in capillary endothelial cells, indicating the activation of a signaling pathway potentially responsible for epidermal abnormalities and contributing to LS pathogenesis. In summary, our scRNA-seq analysis identified potential disease-propagating endothelial cell clusters with upregulated pathways in LS skin, highlighting their importance in disease progression.
Keywords: IL33; IL6; JAG-NOTCH signaling; SELE; XIST; endothelial cells; localized scleroderma; morphea; single-cell RNA sequencing; skin.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Single-Cell Transcriptome Analysis Identifies Subclusters with Inflammatory Fibroblast Responses in Localized Scleroderma.Int J Mol Sci. 2023 Jun 6;24(12):9796. doi: 10.3390/ijms24129796. Int J Mol Sci. 2023. PMID: 37372943 Free PMC article.
-
Transcriptomic Evaluation of Juvenile Localized Scleroderma Skin With Histologic and Clinical Correlation.Arthritis Rheumatol. 2021 Oct;73(10):1921-1930. doi: 10.1002/art.41758. Epub 2021 Aug 31. Arthritis Rheumatol. 2021. PMID: 33844442
-
Spatial Transcriptomics Identifies Cellular and Molecular Characteristics of Scleroderma Skin Lesions: Pilot Study in Juvenile Scleroderma.Int J Mol Sci. 2024 Aug 23;25(17):9182. doi: 10.3390/ijms25179182. Int J Mol Sci. 2024. PMID: 39273131 Free PMC article.
-
Immunopathogenesis of Pediatric Localized Scleroderma.Front Immunol. 2019 Apr 30;10:908. doi: 10.3389/fimmu.2019.00908. eCollection 2019. Front Immunol. 2019. PMID: 31114575 Free PMC article. Review.
-
Cytokine profiles in localized scleroderma and relationship to clinical features.Cytokine. 2011 Aug;55(2):157-64. doi: 10.1016/j.cyto.2011.04.001. Epub 2011 May 4. Cytokine. 2011. PMID: 21536453 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

