Genomic and Transcriptomic Profile of HNF1A-Mutated Liver Adenomas Highlights Molecular Signature and Potential Therapeutic Implications
- PMID: 39408812
- PMCID: PMC11477380
- DOI: 10.3390/ijms251910483
Genomic and Transcriptomic Profile of HNF1A-Mutated Liver Adenomas Highlights Molecular Signature and Potential Therapeutic Implications
Abstract
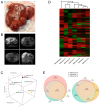
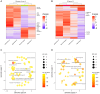
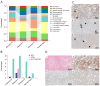
Hepatocellular adenomas (HAs) are tumors that can develop under different conditions, including in patients harboring a germline mutation in HNF1A. However, little is known about the pathogenesis of such disease. This work aims to better define what mechanisms lie under the development of this condition. Six HAs were sampled from the liver of a 17-year-old male affected by diabetes and multiple hepatic adenomatosis harboring the heterozygous pathogenic germline variant c.815G>A, p.(Arg272His) in HNF1A, which has a dominant negative effect. All HAs were molecularly characterized. Four of them were shown to harbor a second somatic HNF1A variant and one had a mutation in the ARID1A gene, while no additional somatic changes were found in the remaining HA and normal parenchyma. A transcriptomic profile of the same HA samples was also performed. HNF1A biallelic mutations were associated with the up-regulation of several pathways including the tricarboxylic acid cycle, the metabolism of fatty acids, and mTOR signaling while angiogenesis, endothelial and vascular proliferation, cell migration/adhesion, and immune response were down-regulated. Contrariwise, in the tumor harboring the ARID1A variant, angiogenesis was up-modulated while fatty acid metabolism was down-modulated. Histological analyses confirmed the molecular data. Independently of the second mutation, energetic processes and cholesterol metabolism were up-modulated, while the immune response was down-modulated. This work provides a complete molecular signature of HNF1A-associated HAs, analyzing the association between specific HNF1A variants and the development of HA while identifying potential new therapeutic targets for non-surgical treatment.
Keywords: HNF1A; genetics; hepatic adenomas; liver; molecular signature.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

