The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer
- PMID: 39408832
- PMCID: PMC11477048
- DOI: 10.3390/ijms251910503
The Use of Patient-Derived Organoids in the Study of Molecular Metabolic Adaptation in Breast Cancer
Abstract
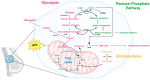
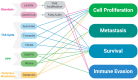
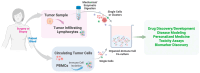
Around 13% of women will likely develop breast cancer during their lifetime. Advances in cancer metabolism research have identified a range of metabolic reprogramming events, such as altered glucose and amino acid uptake, increased reliance on glycolysis, and interactions with the tumor microenvironment (TME), all of which present new opportunities for targeted therapies. However, studying these metabolic networks is challenging in traditional 2D cell cultures, which often fail to replicate the three-dimensional architecture and dynamic interactions of real tumors. To address this, organoid models have emerged as powerful tools. Tumor organoids are 3D cultures, often derived from patient tissue, that more accurately mimic the structural and functional properties of actual tumor tissues in vivo, offering a more realistic model for investigating cancer metabolism. This review explores the unique metabolic adaptations of breast cancer and discusses how organoid models can provide deeper insights into these processes. We evaluate the most advanced tools for studying cancer metabolism in three-dimensional culture models, including optical metabolic imaging (OMI), matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI-MSI), and recent advances in conventional techniques applied to 3D cultures. Finally, we explore the progress made in identifying and targeting potential therapeutic targets in breast cancer metabolism.
Keywords: MALDI-MSI; breast cancer; glycolysis; lipid metabolism; metabolism; optical metabolic imaging; organoid; tumor microenvironment.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- National Cancer Institute Surveillance, Epidemiology, and End Results Program. Breast Cancer—Breast Cancer Risk from Birth over Time, by Sex, All Races/Ethnicities, Risk of Being Diagnosed with Cancer (2018–2019, 2021) [(accessed on 23 August 2024)]; Available online: https://seer.cancer.gov/statistics-network/explorer/
-
- Centers for Disease Control and Prevention. U.S. Department of Health and Human Services U.S. Cancer Statistics Female Breast Cancer Stat Bite. [(accessed on 23 August 2024)];2024 Available online: https://www.cdc.gov/united-states-cancer-statistics/publications/breast-....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

