The SIRT5-Mediated Upregulation of C/EBPβ Promotes White Adipose Tissue Browning by Enhancing UCP1 Signaling
- PMID: 39408844
- PMCID: PMC11476608
- DOI: 10.3390/ijms251910514
The SIRT5-Mediated Upregulation of C/EBPβ Promotes White Adipose Tissue Browning by Enhancing UCP1 Signaling
Abstract
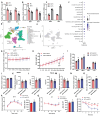
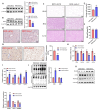
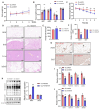
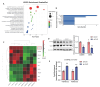
Sirtuin 5 (SIRT5) plays an important role in the maintenance of lipid metabolism and in white adipose tissue browning. In this study, we established a mouse model for diet-induced obesity and the browning of white fat; combined with gene expression intervention, transcriptome sequencing, and cell molecular biology methods, the regulation and molecular mechanisms of SIRT5 on fat deposition and beige fat formation were studied. The results showed that the loss of SIRT5 in obese mice exacerbated white adipose tissue deposition and metabolic inflexibility. Furthermore, the deletion of SIRT5 in a white-fat-browning mouse increased the succinylation of uncoupling protein 1 (UCP1), resulting in a loss of the beiging capacity of the subcutaneous white adipose tissue and impaired cold tolerance. Mechanistically, the inhibition of SIRT5 results in impaired CCAAT/enhancer binding protein beta (C/EBPβ) expression in brown adipocytes, which in turn reduces the UCP1 transcriptional pathway. Thus, the transcription of UCP1 mediated by the SIRT5-C/EBPβ axis is critical in regulating energy balance and obesity-related metabolism.
Keywords: SIRT5; UCP1; fat synthesis; protein succinylation; white adipose tissue browning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

