Gene Expression Aberrations in Alcohol-Associated Hepatocellular Carcinoma
- PMID: 39408891
- PMCID: PMC11476681
- DOI: 10.3390/ijms251910558
Gene Expression Aberrations in Alcohol-Associated Hepatocellular Carcinoma
Abstract
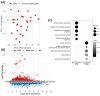
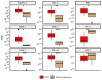
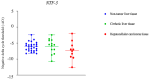
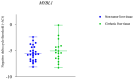
Hepatocellular carcinoma (HCC) is the most prevalent primary liver cancer, ranking as the sixth most common cancer worldwide and the fourth leading cause of cancer-related deaths. Most HCC cases originate from cirrhotic livers, typically due to chronic liver diseases, such as hepatitis B (HBV) and hepatitis C (HCV) infections, and alcoholism. HCC cells often harbor numerous somatic mutations that are implicated in HCC development, but epigenetic factors, such as miRNA interference, can also affect HCC initiation and progress. miRNA-221 has been explored as a factor affecting HCC development in HCC of viral etiology, but little is known about its effects on gene expression in alcohol-associated HCC. This study aimed to explore potentially similar gene expression aberrations underlying viral and alcohol-induced HCC. We analyzed available transcriptome data from non-tumor hepatocytes and viral-induced HCC tissues. The most notable differences in gene expression associated with miRNA-221 between non-tumor hepatocytes and viral-induced HCC involved NTF-3 and MYBL1 genes. To assess these data in alcohol-induced HCC, we examined 111 tissue samples: tumor tissue and cirrhotic tissue samples from 37 HCC patients and 37 samples from non-tumor liver tissue using RT-Q PCR. We found no significant difference in NTF-3 expression, but MYBL1 expression was significantly lower in HCC tissue compared to non-tumor hepatocytes and cirrhotic tissue. Our findings highlight the importance of the MYBL1 gene in HCC development and emphasize the need for diverse approaches in evaluating tumor mechanisms.
Keywords: MYBL1; NTF-3; alcohol etiology; hepatocellular carcinoma; miRNA-221.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
miR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma.World J Gastroenterol. 2016 Jun 14;22(22):5183-92. doi: 10.3748/wjg.v22.i22.5183. World J Gastroenterol. 2016. PMID: 27298561 Free PMC article.
-
Role of microRNA-210-3p in hepatitis B virus-related hepatocellular carcinoma.Am J Physiol Gastrointest Liver Physiol. 2020 Mar 1;318(3):G401-G409. doi: 10.1152/ajpgi.00269.2019. Epub 2020 Jan 6. Am J Physiol Gastrointest Liver Physiol. 2020. PMID: 31905024
-
Viral expression and molecular profiling in liver tissue versus microdissected hepatocytes in hepatitis B virus-associated hepatocellular carcinoma.J Transl Med. 2014 Aug 21;12:230. doi: 10.1186/s12967-014-0230-1. J Transl Med. 2014. PMID: 25141867 Free PMC article.
-
Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases.World J Gastroenterol. 2015 Jun 28;21(24):7375-99. doi: 10.3748/wjg.v21.i24.7375. World J Gastroenterol. 2015. PMID: 26139985 Free PMC article. Review.
-
Conceptual models for the initiation of hepatitis B virus-associated hepatocellular carcinoma.Liver Int. 2015 Jul;35(7):1786-800. doi: 10.1111/liv.12773. Epub 2015 Feb 9. Liver Int. 2015. PMID: 25640596 Review.
Cited by
-
Bioinformatics identification of key microRNA-correlated genes associated with hepatocellular carcinoma heterogeneity and prognosis.BMC Gastroenterol. 2025 Jul 1;25(1):452. doi: 10.1186/s12876-025-04031-6. BMC Gastroenterol. 2025. PMID: 40596892 Free PMC article.
References
-
- World Health Organization (WHO) Who Classification of Tumours: Digestive System Tumours. World Health Organization (WHO); Lyon, France: 2019.
-
- Srivastava A., Allende D.S., Goldblum J.R. Gastrointestinal and Liver Pathology: A Volume in the Series: Foundations in Diagnostic Pathology. Elsevier; Amsterdam, The Netherlands: 2023.
-
- Christopher D.M.F. Diagnostic Histopathology of Tumors. Elsevier; Amsterdam, The Netherlands: 2021.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

