Identification of Grape Laccase Genes and Their Potential Role in Secondary Metabolite Synthesis
- PMID: 39408902
- PMCID: PMC11476532
- DOI: 10.3390/ijms251910574
Identification of Grape Laccase Genes and Their Potential Role in Secondary Metabolite Synthesis
Abstract
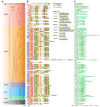
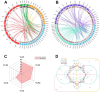
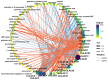
Laccase, a copper-containing oxidoreductase, has close links with secondary metabolite biosynthesis in plants. Its activity can affect the synthesis and accumulation of secondary metabolites, thereby influencing plant growth, development, and stress resistance. This study aims to identify the grape laccases (VviLAC) gene family members in grape (Vitis vinifera L.) and explore the transcriptional regulatory network in berry development. Here, 115 VviLACs were identified and divided into seven (Type I-VII) classes. These were distributed on 17 chromosomes and out of 47 VviLACs on chromosome 18, 34 (72.34%) were involved in tandem duplication events. VviLAC1, VviLAC2, VviLAC3, and VviLAC62 were highly expressed before fruit color development, while VviLAC4, VviLAC12, VviLAC16, VviLAC18, VviLAC20, VviLAC53, VviLAC60 and VviLAC105 were highly expressed after fruit color transformation. Notably, VviLAC105 showed a significant positive correlation with important metabolites including resveratrol, resveratrol dimer, and peonidin-3-glucoside. Analysis of the transcriptional regulatory network predicted that the 12 different transcription factors target VviLACs genes. Specifically, WRKY and ERF were identified as potential transcriptional regulatory factors for VviLAC105, while Dof and MYB were identified as potential transcriptional regulatory factors for VviLAC51. This study identifies and provides basic information on the grape LAC gene family members and, in combination with transcriptome and metabolome data, predicts the upstream transcriptional regulatory network of VviLACs.
Keywords: RNA-seq; berry development; gene family; grapevine; laccase; secondary metabolites.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Zhao Q., Nakashima J., Chen F., Yin Y., Fu C., Yun J., Shao H., Wang X., Wang Z.-Y., Dixon R.A. Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25:3976–3987. doi: 10.1105/tpc.113.117770. - DOI - PMC - PubMed
-
- Iqbal M.J., Ahsan R., Afzal A., Jamai A., Meksem K., El-Shemy H.A., Lightfoot D.A. Multigeneic QTL: The laccase encoded within the soybean Rfs2/rhg1 locus inferred to underlie part of the dual resistance to cyst nematode and sudden death syndrome. Curr. Issues Mol. Biol. 2009;11:I11. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

