Exploring a Potential Optimization Route for Peptide Ligands of the Sam Domain from the Lipid Phosphatase Ship2
- PMID: 39408946
- PMCID: PMC11476629
- DOI: 10.3390/ijms251910616
Exploring a Potential Optimization Route for Peptide Ligands of the Sam Domain from the Lipid Phosphatase Ship2
Abstract
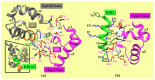
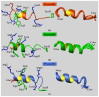
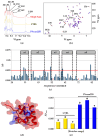
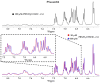
The Sam (Sterile alpha motif) domain of the lipid phosphatase Ship2 (Ship2-Sam) is engaged by the Sam domain of the receptor tyrosine kinase EphA2 (EphA2-Sam) and, this interaction is principally linked to procancer effects. Peptides able to hinder the formation of the EphA2-Sam/Ship2-Sam complex could possess therapeutic potential. Herein, by employing the FoldX software suite, we set up an in silico approach to improve the peptide targeting of the so-called Mid Loop interface of Ship2-Sam, representing the EphA2-Sam binding site. Starting from a formerly identified peptide antagonist of the EphA2-Sam/Ship2-Sam association, first, the most stabilizing mutations that could be inserted in each peptide position were predicted. Then, they were combined, producing a list of potentially enhanced Ship2-Sam ligands. A few of the in silico generated peptides were experimentally evaluated. Interaction assays with Ship2-Sam were performed using NMR and BLI (BioLayer Interferometry). In vitro assays were conducted as well to check for cytotoxic effects against both cancerous and healthy cells, and also to assess the capacity to regulate EphA2 degradation. This study undoubtedly enlarges our knowledge on how to properly target EphA2-Sam/Ship2-Sam associations with peptide-based tools and provides a promising strategy that can be used to target any protein-protein interaction.
Keywords: EphA2 receptor; Sam (Sterile alpha motif) domain; Ship2 lipid phosphatase; anticancer molecular tools; drug design; peptides.
Conflict of interest statement
M.L. and F.A.M. are co-guest editors of the special issue “New Insights into Bioactive Peptides: Design, Synthesis, Structure-Activity Relationship 2.0”.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

