Transcriptomic Analysis of the CNL Gene Family in the Resistant Rice Cultivar IR28 in Response to Ustilaginoidea virens Infection
- PMID: 39408984
- PMCID: PMC11477166
- DOI: 10.3390/ijms251910655
Transcriptomic Analysis of the CNL Gene Family in the Resistant Rice Cultivar IR28 in Response to Ustilaginoidea virens Infection
Abstract
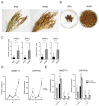
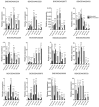
Rice false smut, caused by Ustilaginoidea virens, threatens rice production by reducing yields and contaminating grains with harmful ustiloxins. However, studies on resistance genes are scarce. In this study, the resistance level of IR28 (resistant cultivar) to U. virens was validated through artificial inoculation. Notably, a reactivation of resistance genes after transient down-regulation during the first 3 to 5 dpi was observed in IR28 compared to WX98 (susceptible cultivar). Cluster results of a principal component analysis and hierarchical cluster analysis of differentially expressed genes (DEGs) in the transcriptome exhibited longer expression patterns in the early infection phase of IR28, consistent with its sustained resistance response. Results of GO and KEGG enrichment analyses highlighted the suppression of immune pathways when the hyphae first invade stamen filaments at 5 dpi, but sustained up-regulated DEGs were linked to the 'Plant-pathogen interaction' (osa04626) pathway, notably disease-resistant protein RPM1 (K13457, CNLs, coil-coiled NLR). An analysis of CNLs identified 245 proteins containing Rx-CC and NB-ARC domains in the Oryza sativa Indica genome. Partial candidate CNLs were shown to exhibit up-regulation at both 1 and 5 dpi in IR28. This study provides insights into CNLs' responses to U. virens in IR28, potentially informing resistance mechanisms and genetic breeding targets.
Keywords: CNLs; NLR immune receptor; Ustilaginoidea virens; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Song J.H., Wei W., Lv B., Lin Y., Yin W.X., Peng Y.L., Schnabel G., Huang J.B., Jiang D.H., Luo C.X. Rice false smut fungus hijacks the rice nutrients supply by blocking and mimicking the fertilization of rice ovary. Environ. Microbiol. 2016;18:3840–3849. doi: 10.1111/1462-2920.13343. - DOI - PubMed
-
- Wen Ming W., Jing F., John Martin Jerome J. Rice false smut: An increasing threat to grain yield and quality. In: Yulin J., editor. Protecting Rice Grains in the Post-Genomic Era. IntechOpen; Rijeka, Croatia: 2019. p. Ch.7.
-
- Zhang Y., Xu Q., Sun Q., Kong R., Liu H., Yi X.e., Liang Z., Letcher R.J., Liu C. Ustiloxin a inhibits proliferation of renal tubular epithelial cells in vitro and induces renal injury in mice by disrupting structure and respiratory function of mitochondria. J. Hazard. Mater. 2023;448:130791. doi: 10.1016/j.jhazmat.2023.130791. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

