Proof of Concept for Genome Profiling of the Neurofibroma/Sarcoma Sequence in Neurofibromatosis Type 1
- PMID: 39409151
- PMCID: PMC11476461
- DOI: 10.3390/ijms251910822
Proof of Concept for Genome Profiling of the Neurofibroma/Sarcoma Sequence in Neurofibromatosis Type 1
Abstract
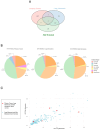
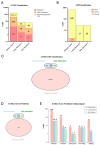
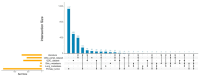
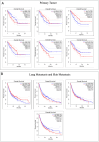
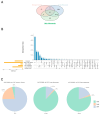
Neurofibromatosis type 1 (NF1) is an autosomal dominant genetic disorder characterized by the predisposition to develop tumors such as malignant peripheral nerve sheath tumors (MPNSTs) which represents the primary cause of death for NF1-affected patients. Regardless of the high incidence and mortality, the molecular mechanisms underneath MPNST growth and metastatic progression remain poorly understood. In this proof-of-concept study, we performed somatic whole-exome sequencing (WES) to profile the genomic alterations in four samples from a patient with NF1-associated MPNST, consisting of a benign plexiform neurofibroma, a primary MPNST, and metastases from lung and skin tissues. By comparing genomic patterns, we identified a high level of variability across samples with distinctive genetic changes which allow for the definition of profiles of the early phase with respect to the late metastatic stages. Pathogenic and likely pathogenic variants were abundant in the primary tumor, whereas the metastatic samples exhibited a high level of copy-number variations (CNVs), highlighting a possible genomic instability in the late phases. The most known MPNST-related genes, such as TP53 and SUZ12, were identified in CNVs observed within the primary tumor. Pathway analysis of altered early genes in MPNST pointed to a potential role in cell motility, division and metabolism. Moreover, we employed survival analysis with the TCGA sarcoma genomic dataset on 262 affected patients, in order to corroborate the predictive significance of the identified early and metastatic MPNST driver genes. Specifically, the expression changes related to the mutated genes, such as in RBMX, PNPLA6 and AGAP2, were associated with reduced patient survival, distinguishing them as potential prognostic biomarkers. This study underlines the relevance of integrating genomic results with clinical information for early diagnosis and prognostic understanding of tumor aggressiveness.
Keywords: MPNST; genomic signature; malignant peripheral nerve sheath tumor; neurofibromatosis type 1; tumor progression; whole exome sequencing (WES).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

