Signature Proteins in Small Extracellular Vesicles of Granulocytes and CD4+ T-Cell Subpopulations Identified by Comparative Proteomic Analysis
- PMID: 39409176
- PMCID: PMC11476868
- DOI: 10.3390/ijms251910848
Signature Proteins in Small Extracellular Vesicles of Granulocytes and CD4+ T-Cell Subpopulations Identified by Comparative Proteomic Analysis
Abstract
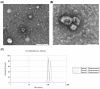
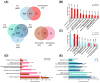
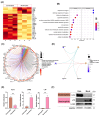
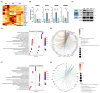
Several studies have described the proteomic profile of different immune cell types, but only a few have also analysed the content of their delivered small extracellular vesicles (sEVs). The aim of the present study was to compare the protein signature of sEVs delivered from granulocytes (i.e., neutrophils and eosinophils) and CD4+ T cells (i.e., TH1, TH2, and TH17) to identify potential biomarkers of the inflammatory profile in chronic inflammatory diseases. Qualitative (DDA) and quantitative (DIA-SWATH) analyses of in vitro-produced sEVs revealed proteome variations depending on the cell source. The main differences were found between granulocyte- and TH cell-derived sEVs, with a higher abundance of antimicrobial proteins (e.g., LCN2, LTF, MPO) in granulocyte-derived sEVs and an enrichment of ribosomal proteins (RPL and RPS proteins) in TH-derived sEVs. Additionally, we found differentially abundant proteins between neutrophil and eosinophil sEVs (e.g., ILF2, LTF, LCN2) and between sEVs from different TH subsets (e.g., ISG15, ITGA4, ITGB2, or NAMPT). A "proof-of-concept" assay was also performed, with TH2 biomarkers ITGA4 and ITGB2 displaying a differential abundance in sEVs from T2high and T2low asthma patients. Thus, our findings highlight the potential use of these sEVs as a source of biomarkers for diseases where the different immune cell subsets studied participate, particularly chronic inflammatory pathologies such as asthma or chronic obstructive pulmonary disease (COPD).
Keywords: exosomes; immune cells; inflammatory diseases; proteomics; small extracellular vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Nguyen D.D., Lai J.-Y. Synthesis, Bioactive Properties, and Biomedical Applications of Intrinsically Therapeutic Nanoparticles for Disease Treatment. Chem. Eng. J. 2022;435:134970. doi: 10.1016/j.cej.2022.134970. - DOI
-
- Vázquez-Mera S., Martelo-Vidal L., Miguéns-Suárez P., Saavedra-Nieves P., Arias P., González-Fernández C., Mosteiro-Añón M., Corbacho-Abelaira M.D., Blanco-Aparicio M., Méndez-Brea P., et al. Serum Exosome Inflamma-miRs Are Surrogate Biomarkers for Asthma Phenotype and Severity. Allergy. 2023;78:141–155. doi: 10.1111/all.15480. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

