Establishment of a Sensitive and Reliable Droplet Digital PCR Assay for the Detection of Bursaphelenchus xylophilus
- PMID: 39409571
- PMCID: PMC11478728
- DOI: 10.3390/plants13192701
Establishment of a Sensitive and Reliable Droplet Digital PCR Assay for the Detection of Bursaphelenchus xylophilus
Abstract
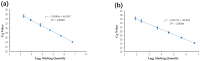
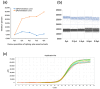
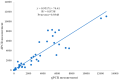
Pine wilt disease (PWD), which poses a significant risk to pine plantations across the globe, is caused by the pathogenic agent Bursaphelenchus xylophilus, also referred to as the pine wood nematode (PWN). A droplet digital PCR (ddPCR) assay was developed for the quick identification of the PWN in order to improve detection sensitivity. The research findings indicate that the ddPCR assay demonstrated significantly higher analysis sensitivity and detection sensitivity in comparison to traditional quantitative PCR (qPCR). However, it had a more limited dynamic range. High specificity was shown by both the ddPCR and qPCR techniques in the diagnosis of the PWN. Assessments of reproducibility revealed that ddPCR had lower coefficients of variation at every template concentration. Inhibition tests showed that ddPCR was less susceptible to inhibitors. There was a strong linear association between standard template measurements obtained using ddPCR and qPCR (Pearson correlation = 0.9317; p < 0.001). Likewise, there was strong agreement (Pearson correlation = 0.9348; p < 0.001) between ddPCR and qPCR measurements in the evaluation of pine wood samples. Additionally, wood samples from symptomatic (100% versus 86.67%) and asymptomatic (31.43% versus 2.9%) pine trees were diagnosed with greater detection rates using ddPCR. This study's conclusions highlight the advantages of the ddPCR assay over qPCR for the quantitative detection of the PWN. This method has a lot of potential for ecological research on PWD and use in quarantines.
Keywords: Bursaphelenchus xylophilus determination; droplet digital PCR; nuclear DNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources

